

| |
||||||||
|
||||||||
Help
Input Fields:
Title:
A job title - any string.PDB files:
You may upload up to 7 pdb files. If your assembly is built of more than 7 components it is necessary to use a stand-alone version of MinkoFit3D. Either a PDB file or a 4 letter PDB code can be used. However, you have to ensure that the downloaded file contains what you want, i.e. downloaded, multimeric structures are treated as they are and are not split into individual chains.Electron density map file:
An electron microscopy electron density map of an assembly in CCP4/MRC formatMap threshold:
An electron density map threshold. If this field is empty, the threshold is automatically calculated.Map resolution:
A resolution of the electron density map.Number of solutions:
A number of alternative solutions you want to be returned by the program.Solution selection method:
Either robust brute-force (checking all possible combinations of individual subunits) or genetic algorithm can be used to select candidate assemblies. When brute force method is selected, 10^5 combination is checked at the maximum. If you have for example 6 subunit complex and choose 10 alternative solutions. Program will use 9 best solutions (according to cross-corelation coefficent). per each subunit to construct final assembly. In case of genetic algorithm all alternative solutions are used to form an initial chromosome.Rotation angle:
An angular sampling step used to generate Euler angles sets.Distribution of Euler angles
Both "lattman" and "uniform" methods avoids degenerate sets of angles. The method proposed by Lattman eliminates degenerated configurations from the equally spaced angles. While the "uniform" distribution is based on random angle generation and polar region avoidance. With the same step number of "lattman" angles is smaller than number of "uniform" angles.Number of map and pdb facets
A number of facets of the meshed electron density map and the pdb structures. If the fields are left empty automatic procedure is used to assign the number of facets of each object. If number of pdb facets is set, all pdb structures are meshed with equal number of facets.Example files
To illustrate the use of MinkoFit3D one can either click "Load Example" button on a submit website or download example files of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil complex (pdb code: 1gte).
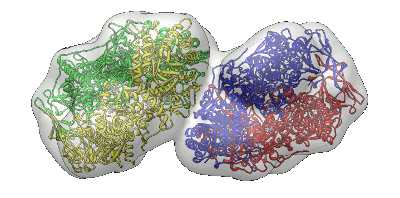
Input:
Title: 1gte
Structures files:
1gte_A.pdb
1gte_B.pdb
1gte_C.pdb
1gte_D.pdb
Electron density map file:
1gte_20.map
Resolution: 20
All other fields might be left with default values.
Output:
There are 2 output files program returns:
1gte_models.zip that contains zipped results.
1gte.txt is a job log file.
|
|

|