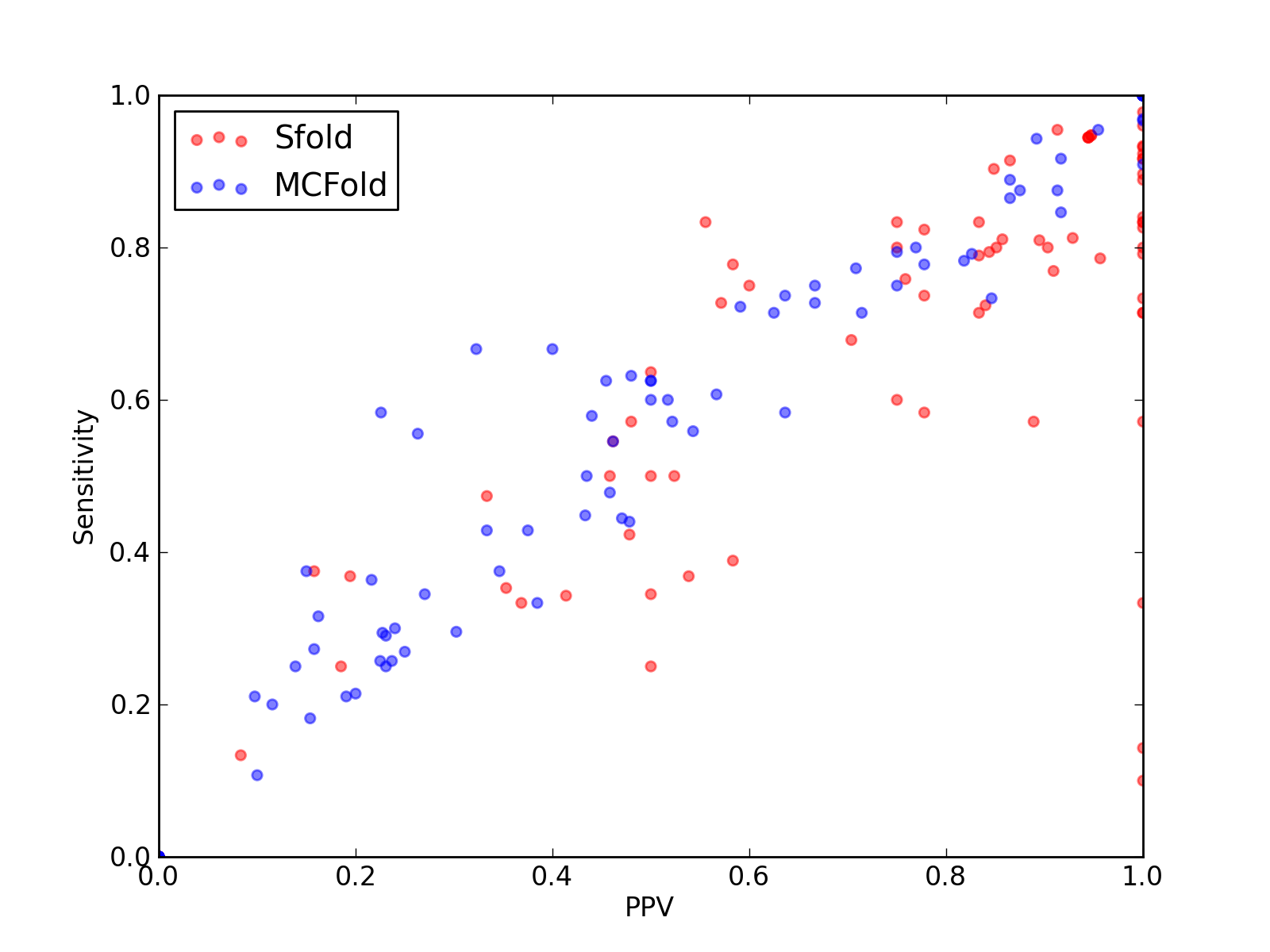
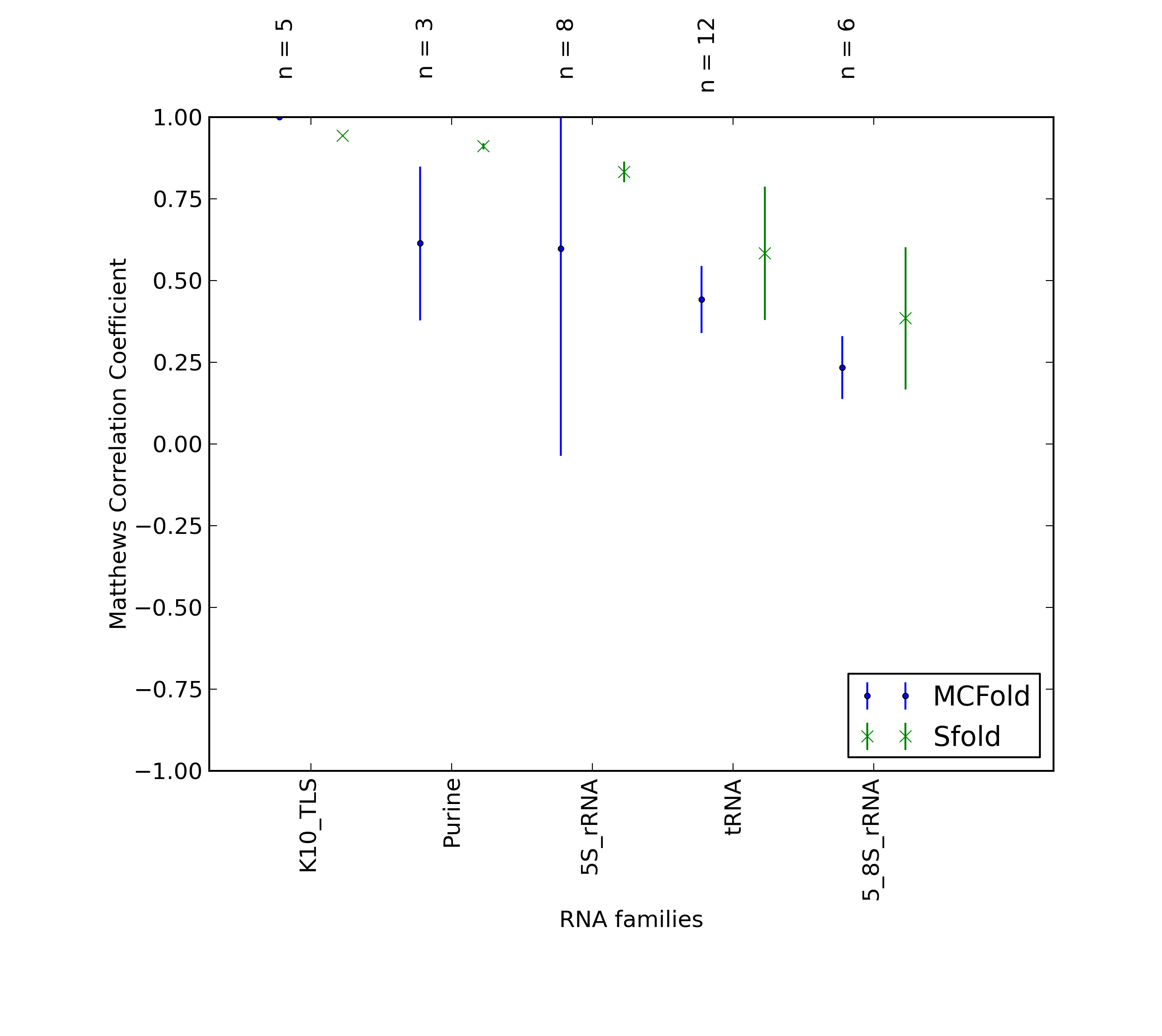
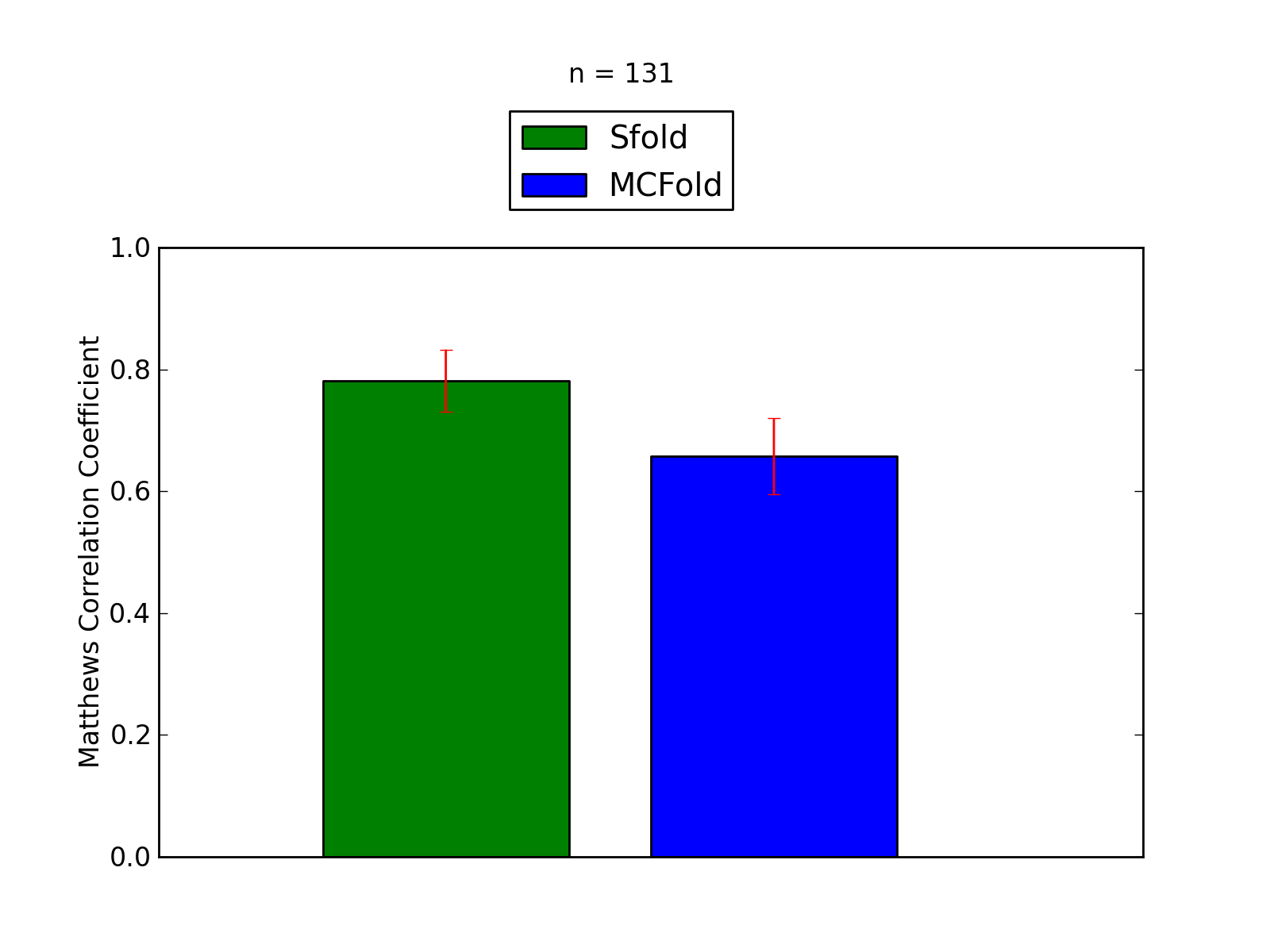
| 2K5Z_A |
-
|
1.00 |
1.00 |
1.00 |
11 |
175 |
0 |
0 |
0 |
0 |
0 |
| 2K63_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
159 |
4 |
0 |
0 |
4 |
0 |
| 2K66_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
80 |
0 |
0 |
0 |
0 |
0 |
| 2KD8_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
99 |
1 |
0 |
0 |
1 |
0 |
| 2KDQ_B |
mir-TAR
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
10 |
164 |
1 |
0 |
0 |
1 |
0 |
| 2KE6_A |
K10_TLS
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
18 |
449 |
1 |
0 |
0 |
1 |
0 |
| 2KEZ_A |
-
|
1.00 |
1.00 |
1.00 |
8 |
94 |
0 |
0 |
0 |
0 |
0 |
| 2KFC_A |
-
|
0.54 |
0.63 |
0.50 |
5 |
226 |
5 |
2 |
3 |
0 |
3 |
| 2KGP_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
112 |
1 |
0 |
0 |
1 |
0 |
| 2KMJ_A |
-
|
1.00 |
1.00 |
1.00 |
11 |
157 |
1 |
0 |
0 |
1 |
0 |
| 2KPV_A |
-
|
1.00 |
1.00 |
1.00 |
13 |
200 |
0 |
0 |
0 |
0 |
0 |
| 2KU0_A |
-
|
1.00 |
1.00 |
1.00 |
12 |
280 |
1 |
0 |
0 |
1 |
0 |
| 2KUR_A |
K10_TLS
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
19 |
448 |
0 |
0 |
0 |
0 |
0 |
| 2KUU_A |
K10_TLS
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
18 |
429 |
1 |
0 |
0 |
1 |
0 |
| 2KUV_A |
K10_TLS
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
19 |
420 |
0 |
0 |
0 |
0 |
0 |
| 2KUW_A |
K10_TLS
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
18 |
452 |
1 |
0 |
0 |
1 |
0 |
| 2KX8_A |
7SK
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
16 |
355 |
0 |
0 |
0 |
0 |
0 |
| 2KXM_A |
-
|
1.00 |
1.00 |
1.00 |
8 |
141 |
1 |
0 |
0 |
1 |
0 |
| 2KZL_A |
-
|
1.00 |
1.00 |
1.00 |
13 |
507 |
5 |
0 |
0 |
5 |
0 |
| 2L1F_A |
Gammaretro_CES
links to Rfam database...
|
0.79 |
0.78 |
0.82 |
18 |
741 |
4 |
0 |
4 |
0 |
5 |
| 2L1F_B |
Gammaretro_CES
links to Rfam database...
|
0.80 |
0.79 |
0.83 |
19 |
768 |
4 |
0 |
4 |
0 |
5 |
| 2L2K_A |
-
|
1.00 |
1.00 |
1.00 |
17 |
332 |
1 |
0 |
0 |
1 |
0 |
| 2L3C_B |
-
|
1.00 |
1.00 |
1.00 |
14 |
225 |
1 |
0 |
0 |
1 |
0 |
| 2L3E_A |
-
|
1.00 |
1.00 |
1.00 |
12 |
228 |
2 |
0 |
0 |
2 |
0 |
| 2L3J_B |
mir-2985-2
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
30 |
955 |
0 |
0 |
0 |
0 |
0 |
| 2L5Z_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
102 |
0 |
0 |
0 |
0 |
0 |
| 2L94_A |
HIV_FE
links to Rfam database...
|
1.00 |
1.00 |
1.00 |
18 |
339 |
1 |
0 |
0 |
1 |
0 |
| 2LA5_A |
-
|
-0.03 |
0.00 |
0.00 |
0 |
265 |
10 |
0 |
10 |
0 |
7 |
| 2LBS_A |
-
|
1.00 |
1.00 |
1.00 |
14 |
193 |
1 |
0 |
0 |
1 |
0 |
| 2LC8_A |
GP_knot1
links to Rfam database...
|
0.44 |
0.44 |
0.47 |
8 |
511 |
10 |
0 |
9 |
1 |
10 |
| 2LDL_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
131 |
1 |
0 |
0 |
1 |
0 |
| 2LDT_A |
-
|
0.95 |
0.91 |
1.00 |
10 |
152 |
0 |
0 |
0 |
0 |
1 |
| 2LHP_A |
-
|
1.00 |
1.00 |
1.00 |
15 |
246 |
1 |
0 |
0 |
1 |
0 |
| 2LI4_A |
-
|
1.00 |
1.00 |
1.00 |
14 |
175 |
0 |
0 |
0 |
0 |
0 |
| 2LJJ_A |
-
|
1.00 |
1.00 |
1.00 |
7 |
123 |
3 |
0 |
0 |
3 |
0 |
| 2LK3_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
91 |
0 |
0 |
0 |
0 |
0 |
| 2LKR_A |
-
|
1.00 |
1.00 |
1.00 |
29 |
2411 |
11 |
0 |
0 |
11 |
0 |
| 2LQZ_A |
-
|
1.00 |
1.00 |
1.00 |
8 |
124 |
3 |
0 |
0 |
3 |
0 |
| 2LU0_A |
-
|
1.00 |
1.00 |
1.00 |
16 |
422 |
2 |
0 |
0 |
2 |
0 |
| 2LWK_A |
-
|
1.00 |
1.00 |
1.00 |
11 |
196 |
2 |
0 |
0 |
2 |
0 |
| 2M58_A |
-
|
0.22 |
0.25 |
0.23 |
3 |
531 |
13 |
1 |
9 |
3 |
9 |
| 2RP0_A |
-
|
0.70 |
0.71 |
0.71 |
5 |
109 |
2 |
1 |
1 |
0 |
2 |
| 2RPT_A |
-
|
1.00 |
1.00 |
1.00 |
7 |
65 |
0 |
0 |
0 |
0 |
0 |
| 2RRC_A |
-
|
1.00 |
1.00 |
1.00 |
7 |
68 |
1 |
0 |
0 |
1 |
0 |
| 2WRQ_Y |
tRNA
links to Rfam database...
|
0.38 |
0.56 |
0.26 |
5 |
1133 |
21 |
8 |
6 |
7 |
4 |
| 2WW9_F |
-
|
0.68 |
0.75 |
0.67 |
6 |
103 |
4 |
1 |
2 |
1 |
2 |
| 2WW9_D |
-
|
-0.02 |
0.00 |
0.00 |
0 |
723 |
21 |
6 |
12 |
3 |
10 |
| 2WW9_E |
-
|
-0.03 |
0.00 |
0.00 |
0 |
165 |
8 |
1 |
6 |
1 |
5 |
| 2WWQ_V |
tRNA
links to Rfam database...
|
0.19 |
0.21 |
0.19 |
4 |
1183 |
20 |
3 |
14 |
3 |
15 |
| 2XKV_B |
Bacteria_small_SRP
links to Rfam database...
|
0.20 |
0.27 |
0.16 |
3 |
1816 |
30 |
4 |
12 |
14 |
8 |
| 2XQD_Y |
tRNA
links to Rfam database...
|
0.39 |
0.43 |
0.38 |
9 |
1105 |
18 |
2 |
13 |
3 |
12 |
| 2XXA_G |
Bacteria_small_SRP
links to Rfam database...
|
0.23 |
0.26 |
0.24 |
9 |
2007 |
30 |
1 |
28 |
1 |
26 |
| 2Y8W_B |
-
|
1.00 |
1.00 |
1.00 |
6 |
82 |
2 |
0 |
0 |
2 |
0 |
| 2Y9C_V |
-
|
0.65 |
0.72 |
0.59 |
13 |
884 |
12 |
1 |
8 |
3 |
5 |
| 2YIE_Z |
-
|
0.53 |
0.63 |
0.45 |
5 |
591 |
8 |
2 |
4 |
2 |
3 |
| 2YIE_X |
-
|
-0.01 |
0.00 |
0.00 |
0 |
535 |
15 |
1 |
8 |
6 |
7 |
| 2ZY6_A |
-
|
0.77 |
0.78 |
0.78 |
7 |
267 |
6 |
1 |
1 |
4 |
2 |
| 2ZZN_D |
tRNA
links to Rfam database...
|
0.69 |
0.73 |
0.67 |
16 |
960 |
11 |
1 |
7 |
3 |
6 |
| 3A2K_C |
tRNA
links to Rfam database...
|
0.49 |
0.55 |
0.46 |
12 |
1082 |
14 |
3 |
11 |
0 |
10 |
| 3A3A_A |
tRNA-Sec
links to Rfam database...
|
0.98 |
0.97 |
1.00 |
29 |
1471 |
0 |
0 |
0 |
0 |
1 |
| 3AKZ_H |
tRNA
links to Rfam database...
|
0.46 |
0.50 |
0.43 |
10 |
1104 |
16 |
5 |
8 |
3 |
10 |
| 3AM1_B |
-
|
1.00 |
1.00 |
1.00 |
29 |
1437 |
1 |
0 |
0 |
1 |
0 |
| 3AMU_B |
tRNA
links to Rfam database...
|
0.50 |
0.58 |
0.44 |
11 |
1132 |
15 |
4 |
10 |
1 |
8 |
| 3DW4_A |
-
|
1.00 |
1.00 |
1.00 |
6 |
134 |
3 |
0 |
0 |
3 |
0 |
| 3GCA_A |
-
|
-0.05 |
0.00 |
0.00 |
0 |
150 |
8 |
4 |
4 |
0 |
7 |
| 3GX2_A |
SAM
links to Rfam database...
|
0.58 |
0.61 |
0.57 |
17 |
1419 |
15 |
1 |
12 |
2 |
11 |
| 3HAY_E |
-
|
1.00 |
1.00 |
1.00 |
14 |
910 |
10 |
0 |
0 |
10 |
0 |
| 3HJW_D |
-
|
1.00 |
1.00 |
1.00 |
16 |
593 |
5 |
0 |
0 |
5 |
0 |
| 3IAB_R |
-
|
1.00 |
1.00 |
1.00 |
12 |
373 |
4 |
0 |
0 |
4 |
0 |
| 3ID5_D |
-
|
0.54 |
0.60 |
0.50 |
3 |
223 |
7 |
1 |
2 |
4 |
2 |
| 3IVN_B |
Purine
links to Rfam database...
|
0.45 |
0.48 |
0.46 |
11 |
879 |
14 |
5 |
8 |
1 |
12 |
| 3IWN_A |
c-di-GMP-I
links to Rfam database...
|
0.19 |
0.21 |
0.20 |
6 |
1442 |
24 |
5 |
19 |
0 |
22 |
| 3IZF_C |
5S_rRNA
links to Rfam database...
|
0.92 |
0.94 |
0.89 |
33 |
2603 |
10 |
0 |
4 |
6 |
2 |
| 3J0L_8 |
-
|
1.00 |
1.00 |
1.00 |
7 |
69 |
0 |
0 |
0 |
0 |
0 |
| 3J0L_2 |
-
|
0.25 |
0.27 |
0.25 |
7 |
2222 |
29 |
4 |
17 |
8 |
19 |
| 3J0L_g |
-
|
-0.02 |
0.00 |
0.00 |
0 |
170 |
7 |
4 |
2 |
1 |
2 |
| 3J0L_7 |
-
|
-0.02 |
0.00 |
0.00 |
0 |
504 |
15 |
6 |
9 |
0 |
10 |
| 3J0L_h |
-
|
0.98 |
0.97 |
1.00 |
31 |
2109 |
2 |
0 |
0 |
2 |
1 |
| 3J0L_1 |
-
|
0.88 |
0.85 |
0.92 |
11 |
472 |
5 |
0 |
1 |
4 |
2 |
| 3J0L_a |
-
|
0.14 |
0.18 |
0.15 |
2 |
398 |
13 |
1 |
10 |
2 |
9 |
| 3J16_L |
tRNA
links to Rfam database...
|
0.54 |
0.57 |
0.52 |
12 |
1136 |
12 |
4 |
7 |
1 |
9 |
| 3J20_0 |
tRNA
links to Rfam database...
|
0.66 |
0.71 |
0.63 |
15 |
1195 |
11 |
3 |
6 |
2 |
6 |
| 3J2L_3 |
5S_rRNA
links to Rfam database...
|
0.77 |
0.79 |
0.75 |
27 |
2984 |
15 |
1 |
8 |
6 |
7 |
| 3J3D_C |
tRNA
links to Rfam database...
|
0.54 |
0.63 |
0.48 |
12 |
943 |
13 |
4 |
9 |
0 |
7 |
| 3J3E_8 |
5_8S_rRNA
links to Rfam database...
|
0.15 |
0.20 |
0.12 |
3 |
2716 |
36 |
11 |
12 |
13 |
12 |
| 3J3E_7 |
5S_rRNA
links to Rfam database...
|
0.55 |
0.56 |
0.54 |
19 |
2706 |
23 |
1 |
15 |
7 |
15 |
| 3J3F_7 |
5S_rRNA
links to Rfam database...
|
0.88 |
0.89 |
0.86 |
32 |
2897 |
9 |
1 |
4 |
4 |
4 |
| 3J3F_8 |
5_8S_rRNA
links to Rfam database...
|
0.14 |
0.21 |
0.10 |
4 |
4720 |
51 |
17 |
20 |
14 |
15 |
| 3JYV_7 |
tRNA
links to Rfam database...
|
0.25 |
0.30 |
0.24 |
6 |
1086 |
20 |
8 |
11 |
1 |
14 |
| 3JYX_3 |
5S_rRNA
links to Rfam database...
|
0.46 |
0.67 |
0.32 |
10 |
2347 |
28 |
15 |
6 |
7 |
5 |
| 3JYX_4 |
5_8S_rRNA
links to Rfam database...
|
0.36 |
0.58 |
0.23 |
7 |
4725 |
37 |
19 |
5 |
13 |
5 |
| 3KTW_C |
-
|
0.55 |
0.60 |
0.52 |
15 |
1751 |
20 |
4 |
10 |
6 |
10 |
| 3LA5_A |
Purine
links to Rfam database...
|
0.44 |
0.44 |
0.48 |
11 |
931 |
12 |
2 |
10 |
0 |
14 |
| 3NDB_M |
-
|
0.29 |
0.30 |
0.30 |
13 |
3671 |
35 |
1 |
29 |
5 |
31 |
| 3NKB_B |
-
|
0.68 |
0.74 |
0.64 |
14 |
713 |
8 |
2 |
6 |
0 |
5 |
| 3NMU_E |
-
|
0.51 |
0.67 |
0.40 |
2 |
211 |
8 |
1 |
2 |
5 |
1 |
| 3O58_3 |
5_8S_rRNA
links to Rfam database...
|
0.28 |
0.36 |
0.22 |
8 |
4727 |
39 |
14 |
15 |
10 |
14 |
| 3O58_2 |
5S_rRNA
links to Rfam database...
|
0.25 |
0.29 |
0.23 |
9 |
2715 |
31 |
6 |
24 |
1 |
22 |
| 3OVB_D |
-
|
1.00 |
1.00 |
1.00 |
11 |
215 |
1 |
0 |
0 |
1 |
0 |
| 3OVS_D |
-
|
1.00 |
1.00 |
1.00 |
12 |
204 |
0 |
0 |
0 |
0 |
0 |
| 3P22_G |
-
|
1.00 |
1.00 |
1.00 |
11 |
301 |
1 |
0 |
0 |
1 |
0 |
| 3PDR_A |
ykoK
links to Rfam database...
|
0.78 |
0.80 |
0.77 |
40 |
4788 |
14 |
5 |
7 |
2 |
10 |
| 3R4F_A |
-
|
0.95 |
0.95 |
0.95 |
21 |
884 |
4 |
0 |
1 |
3 |
1 |
| 3R9X_C |
-
|
0.87 |
0.88 |
0.88 |
7 |
221 |
5 |
0 |
1 |
4 |
1 |
| 3RKF_A |
Purine
links to Rfam database...
|
0.89 |
0.88 |
0.91 |
21 |
843 |
3 |
0 |
2 |
1 |
3 |
| 3SD1_A |
THF
links to Rfam database...
|
0.43 |
0.45 |
0.43 |
13 |
1503 |
17 |
1 |
16 |
0 |
16 |
| 3SIU_F |
-
|
0.53 |
0.63 |
0.50 |
5 |
135 |
5 |
2 |
3 |
0 |
3 |
| 3SN2_B |
IRE_II
links to Rfam database...
|
0.58 |
0.58 |
0.64 |
7 |
143 |
4 |
0 |
4 |
0 |
5 |
| 3TRZ_Z |
-
|
-0.05 |
0.00 |
0.00 |
0 |
88 |
4 |
0 |
4 |
0 |
5 |
| 3TS0_U |
-
|
1.00 |
1.00 |
1.00 |
6 |
112 |
1 |
0 |
0 |
1 |
0 |
| 3TS2_V |
-
|
-0.05 |
0.00 |
0.00 |
0 |
102 |
7 |
0 |
6 |
1 |
5 |
| 3U4M_B |
-
|
0.74 |
0.77 |
0.71 |
17 |
1252 |
10 |
1 |
6 |
3 |
5 |
| 3VJR_D |
-
|
1.00 |
1.00 |
1.00 |
12 |
239 |
1 |
0 |
0 |
1 |
0 |
| 3ZEX_G |
-
|
0.00 |
0.00 |
0.00 |
0 |
6493 |
14 |
6 |
8 |
0 |
45 |
| 3ZEX_F |
-
|
0.00 |
0.00 |
0.00 |
0 |
909 |
22 |
0 |
5 |
17 |
4 |
| 3ZEX_C |
5_8S_rRNA
links to Rfam database...
|
0.30 |
0.34 |
0.27 |
10 |
5337 |
28 |
6 |
21 |
1 |
19 |
| 3ZEX_H |
-
|
0.22 |
0.32 |
0.16 |
6 |
3588 |
36 |
15 |
16 |
5 |
13 |
| 3ZEX_D |
5S_rRNA
links to Rfam database...
|
0.23 |
0.26 |
0.23 |
9 |
2756 |
33 |
6 |
25 |
2 |
26 |
| 3ZND_W |
tRNA
links to Rfam database...
|
0.23 |
0.38 |
0.15 |
3 |
1171 |
26 |
9 |
8 |
9 |
5 |
| 4A1C_2 |
5_8S_rRNA
links to Rfam database...
|
0.18 |
0.25 |
0.14 |
5 |
4480 |
45 |
12 |
19 |
14 |
15 |
| 4A1C_3 |
5S_rRNA
links to Rfam database...
|
0.86 |
0.86 |
0.86 |
32 |
2726 |
9 |
0 |
5 |
4 |
5 |
| 4A4U_A |
-
|
1.00 |
1.00 |
1.00 |
9 |
96 |
0 |
0 |
0 |
0 |
0 |
| 4ATO_G |
-
|
0.36 |
0.43 |
0.33 |
3 |
211 |
8 |
0 |
6 |
2 |
4 |
| 4ENB_A |
crcB
links to Rfam database...
|
0.78 |
0.73 |
0.85 |
11 |
459 |
4 |
0 |
2 |
2 |
4 |
| 4ENC_A |
crcB
links to Rfam database...
|
0.34 |
0.33 |
0.38 |
5 |
483 |
11 |
0 |
8 |
3 |
10 |
| 4FNJ_A |
-
|
0.91 |
0.92 |
0.92 |
11 |
238 |
1 |
0 |
1 |
0 |
1 |
| 4FRG_B |
AdoCbl-variant
links to Rfam database...
|
0.35 |
0.38 |
0.35 |
9 |
1176 |
17 |
3 |
14 |
0 |
15 |
| 4FRN_A |
AdoCbl-variant
links to Rfam database...
|
0.09 |
0.11 |
0.10 |
3 |
1818 |
28 |
2 |
25 |
1 |
25 |
| 4HXH_A |
-
|
1.00 |
1.00 |
1.00 |
6 |
89 |
0 |
0 |
0 |
0 |
0 |
| 4JF2_A |
preQ1-II
links to Rfam database...
|
0.74 |
0.75 |
0.75 |
18 |
1058 |
7 |
1 |
5 |
1 |
6 |
| 4JRC_A |
-
|
0.24 |
0.29 |
0.23 |
5 |
600 |
17 |
0 |
17 |
0 |
12 |