Table of contents:
- Overview
- Performance Plots
- Performance of PETfold_pre2.0(seed)
- scored higher in this pairwise comparison
- Performance of MXScarna(seed)
- scored lower in this pairwise comparison
- Compile and download dataset for PETfold_pre2.0(seed) & MXScarna(seed) [.zip] - may take several seconds...
Overview
| Metric |
PETfold_pre2.0(seed) |
|
MXScarna(seed) |
|---|
| MCC |
0.713 |
>
|
0.632 |
| Average MCC ± 95% Confidence Intervals |
0.694
±
0.061
|
>
|
0.572
±
0.093
|
| Sensitivity |
0.600 |
>
|
0.525 |
| Positive Predictive Value |
0.848 |
>
|
0.763 |
| Total TP |
915 |
>
|
800 |
| Total TN |
1248931 |
<
|
1248961 |
| Total FP |
201 |
<
|
304 |
| Total FP CONTRA |
18 |
<
|
34 |
| Total FP INCONS |
146 |
<
|
215 |
| Total FP COMP |
37 |
<
|
55 |
| Total FN |
609 |
<
|
724 |
| P-value |
5.02343278931e-08 |
Performance plots
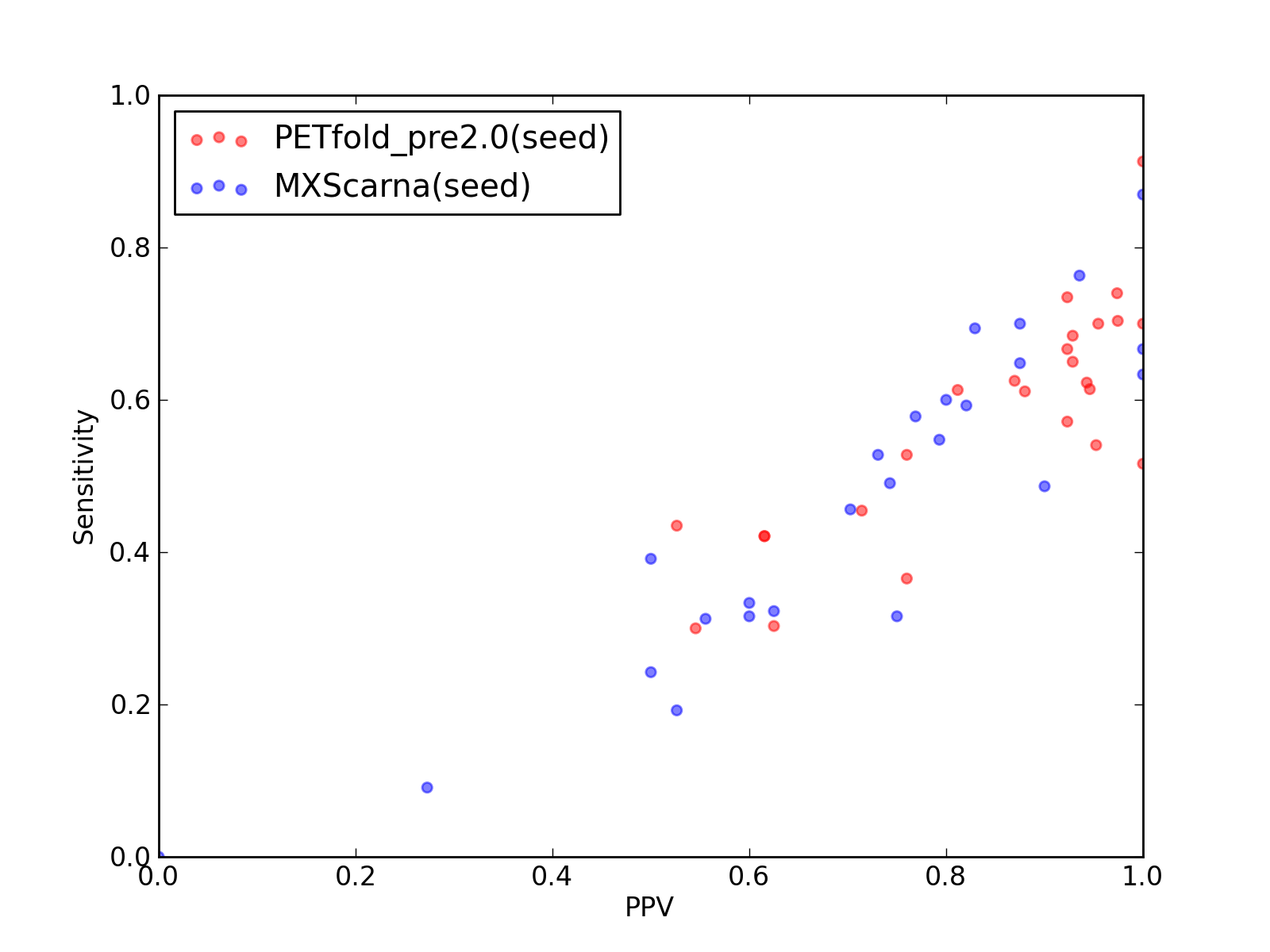
-
Comparison of performance of PETfold_pre2.0(seed) and MXScarna(seed). Positive Predictive Value (PPV) is plotted against sensitivity. Each dot represents a single test of each method. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
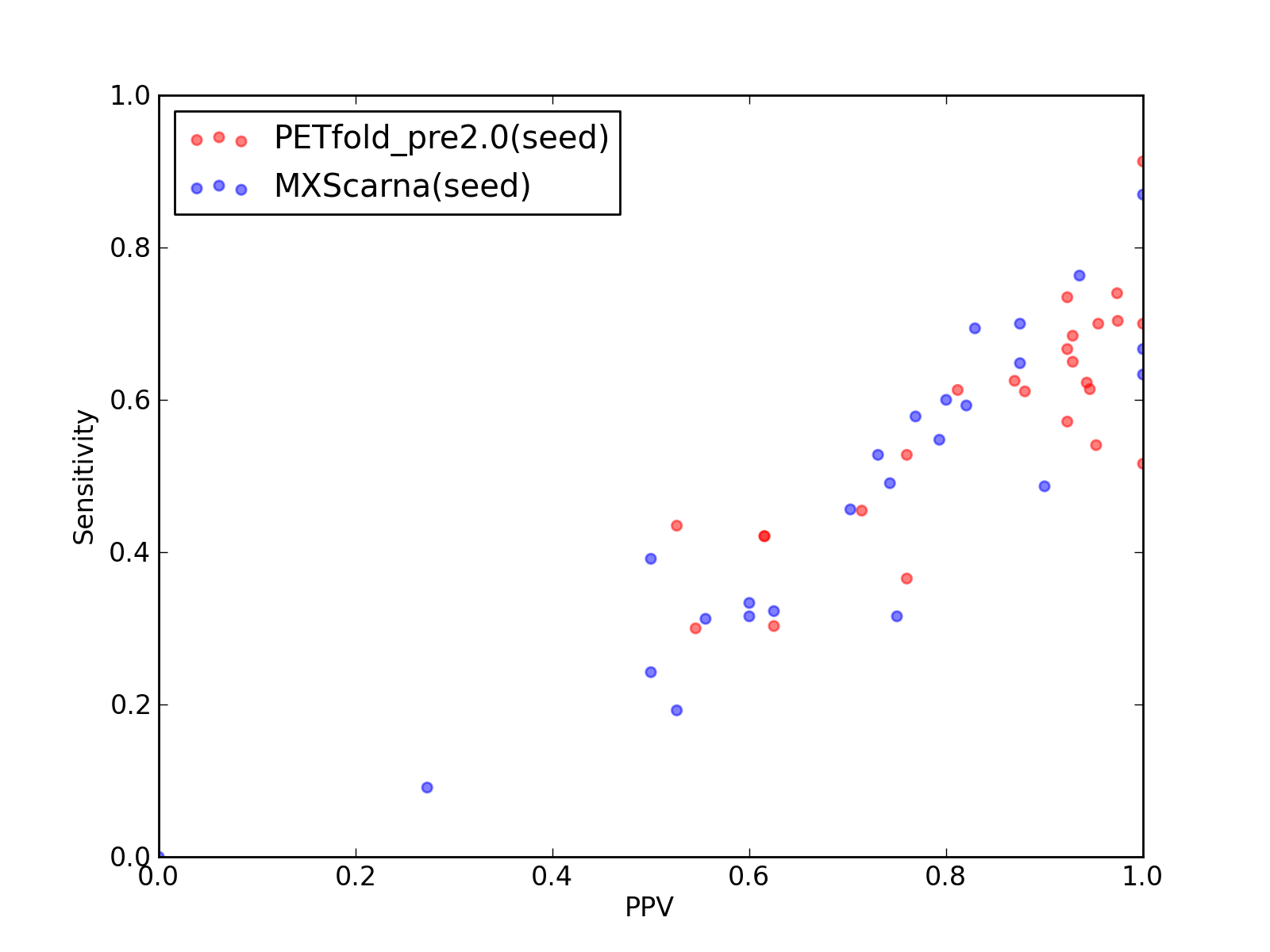
-
Comparison of performance of PETfold_pre2.0(seed) and MXScarna(seed). Positive Predictive Value (PPV) is plotted against sensitivity. Each dot represents a single test of each method. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
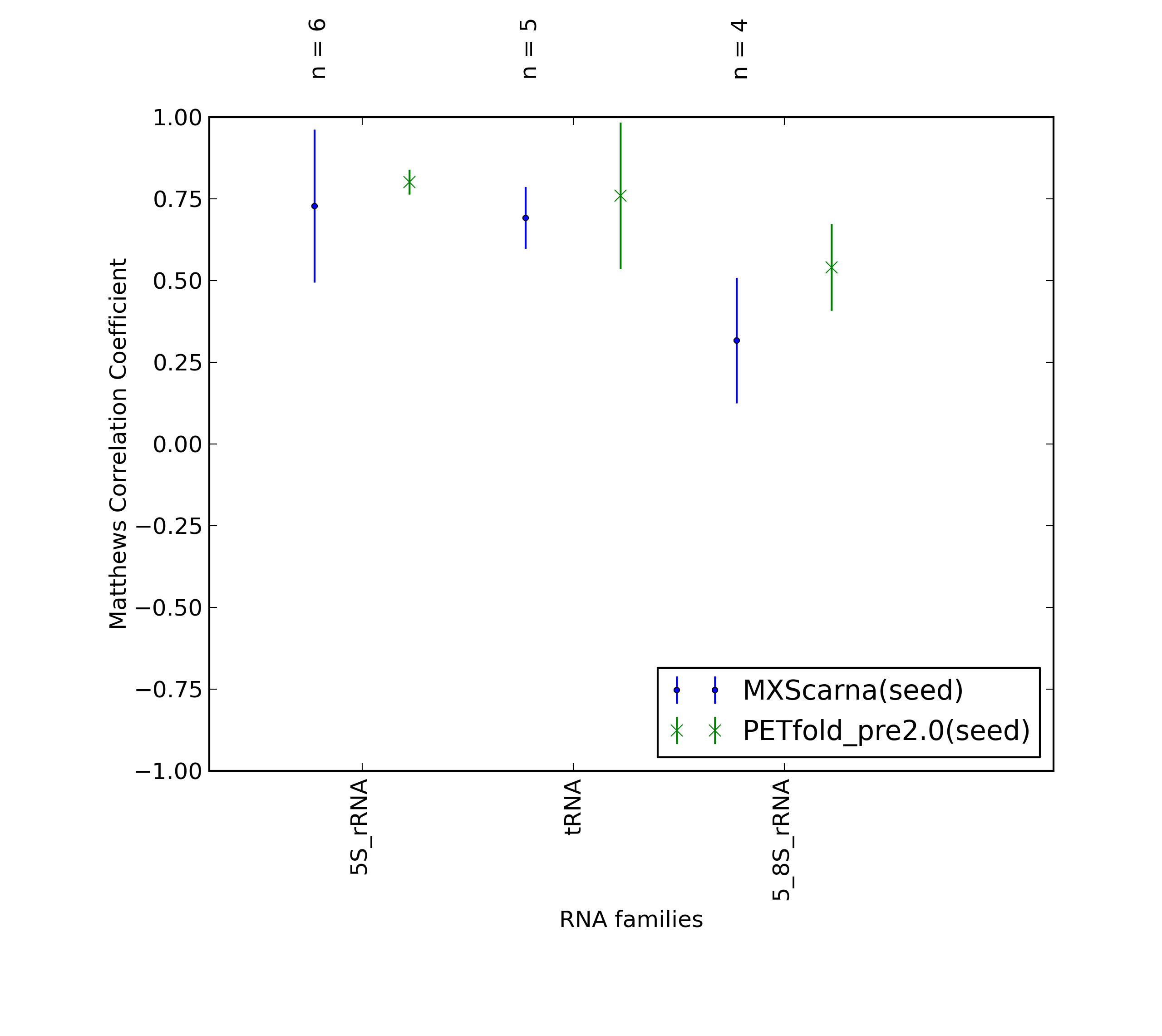
-
Average Matthews Correlation Coefficients (MCC) with 95% confidence intervals (CIs) were plotted for different RNA families, for which at least 3 members were present in the benchmarking dataset. 'n' denotes the number of MCCs used to calculate the average and CI. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
-
Average Matthews Correlation Coefficients (MCC) with 95% confidence intervals (CIs) were plotted for different RNA families, for which at least 3 members were present in the benchmarking dataset. 'n' denotes the number of MCCs used to calculate the average and CI. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
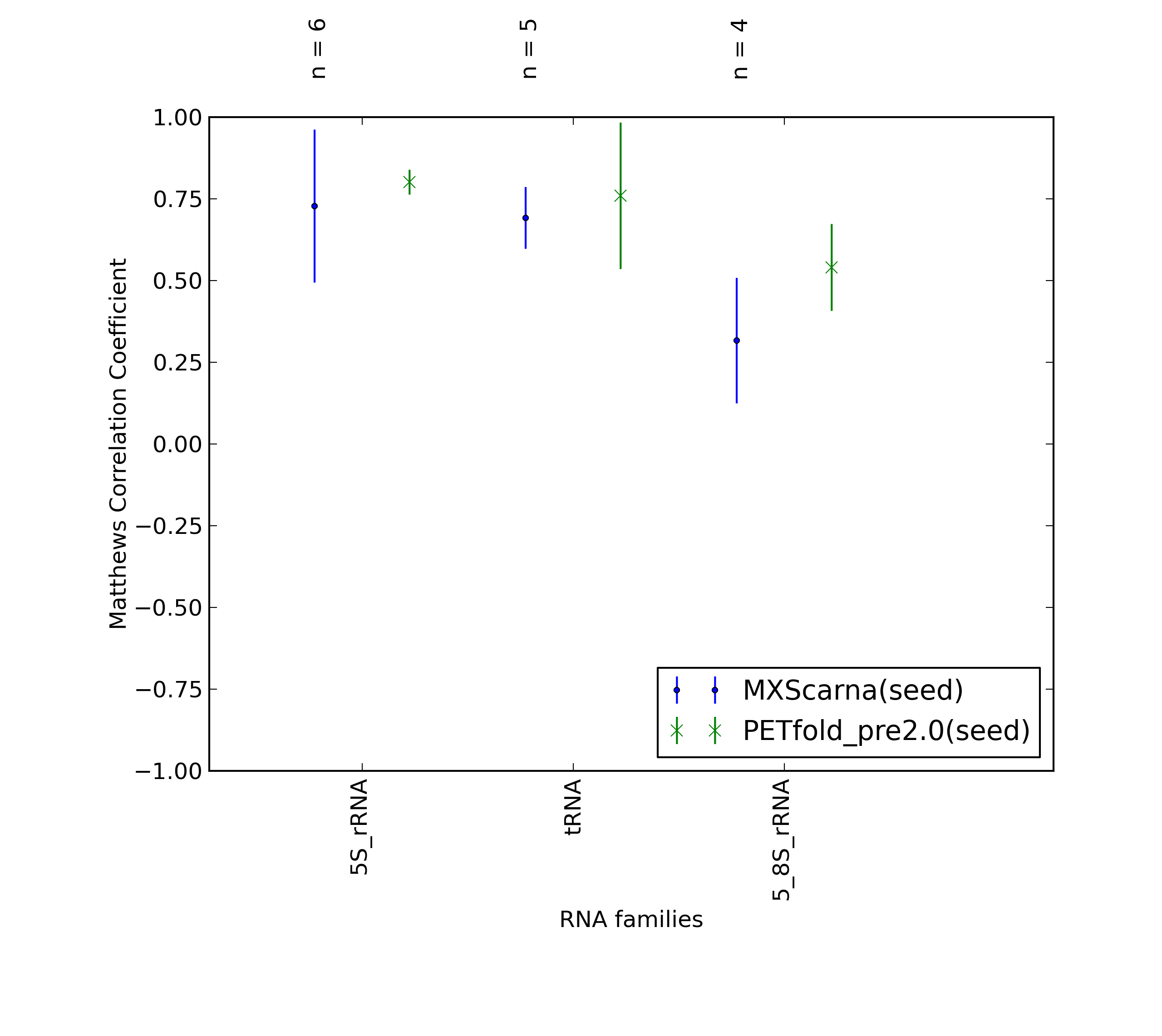
-
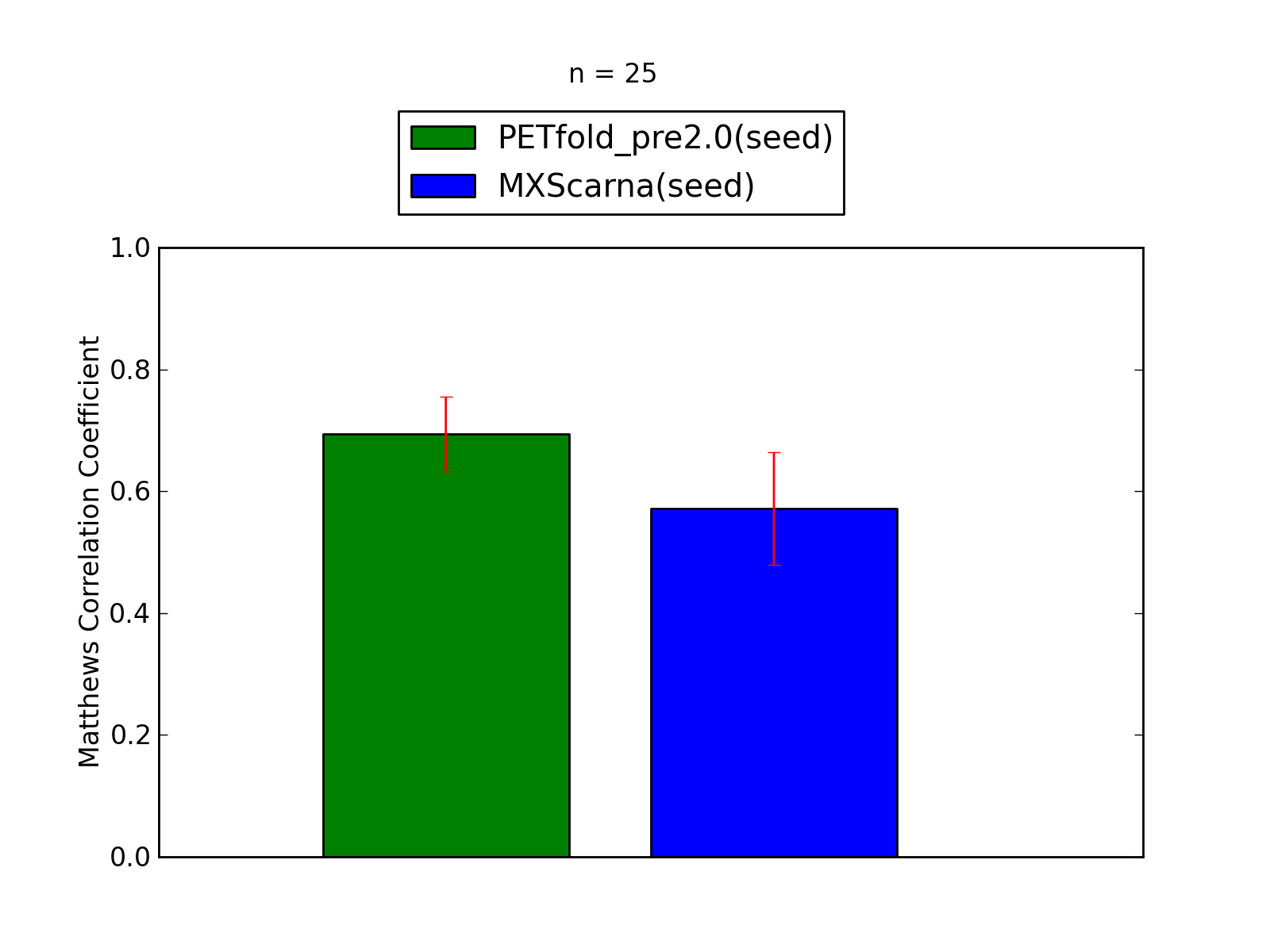
Comparison of average Matthews Correlation Coefficients (MCCs) for PETfold_pre2.0(seed) and MXScarna(seed). The whiskers correspond to 95% confidence intervals (CIs). 'n' denotes the number of MCCs used to calculate average MCCs and CIs. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
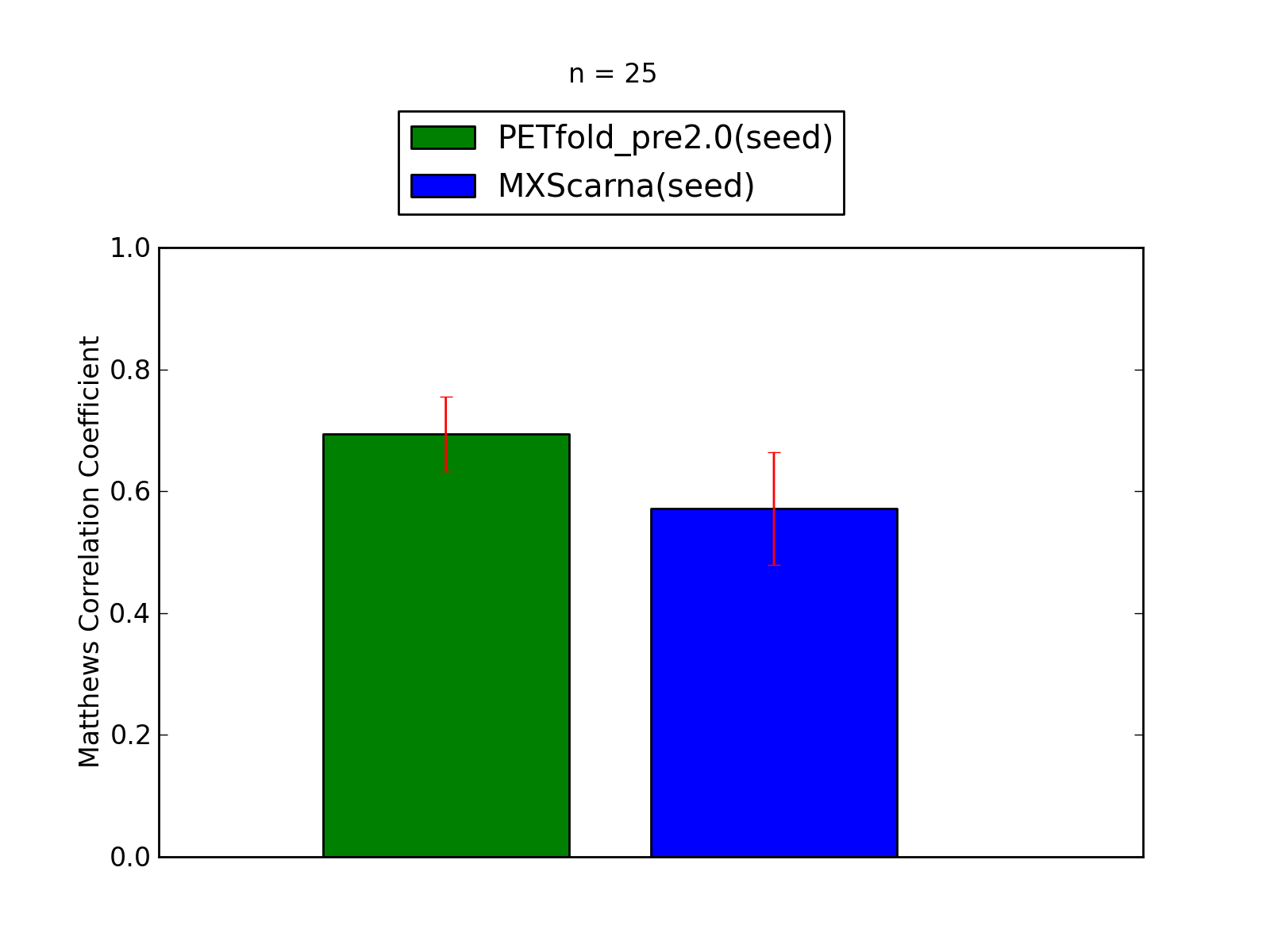
-
Comparison of average Matthews Correlation Coefficients (MCCs) for PETfold_pre2.0(seed) and MXScarna(seed). The whiskers correspond to 95% confidence intervals (CIs). 'n' denotes the number of MCCs used to calculate average MCCs and CIs. See tables below for raw data
(individual counts for PETfold_pre2.0(seed)
and MXScarna(seed)).
Performance of PETfold_pre2.0(seed)
- scored higher in this pairwise comparison
1. Total counts & total scores for PETfold_pre2.0(seed)
| Total Base Pair Counts |
|---|
| Total TP |
915 |
| Total TN |
1248931 |
| Total FP |
201 |
| Total FP CONTRA |
18 |
| Total FP INCONS |
146 |
| Total FP COMP |
37 |
| Total FN |
609 |
| Total Scores |
|---|
| MCC |
0.713 |
| Average MCC ± 95% Confidence Intervals |
0.694
±
0.061
|
| Sensitivity |
0.600 |
| Positive Predictive Value |
0.848 |
| Nr of predictions |
25 |
Performance of MXScarna(seed)
- scored lower in this pairwise comparison
1. Total counts & total scores for MXScarna(seed)
| Total Base Pair Counts |
|---|
| Total TP |
800 |
| Total TN |
1248961 |
| Total FP |
304 |
| Total FP CONTRA |
34 |
| Total FP INCONS |
215 |
| Total FP COMP |
55 |
| Total FN |
724 |
| Total Scores |
|---|
| MCC |
0.632 |
| Average MCC ± 95% Confidence Intervals |
0.572
±
0.093
|
| Sensitivity |
0.525 |
| Positive Predictive Value |
0.763 |
| Nr of predictions |
25 |
Matthews Correlation Coeffient, Sensitivity and Positive Predictive Value have been calculated based
on the paper by Gardener & Giegerich, 2004.
| 






