Table of contents:
- Overview
- Performance Plots
- Performance of RNASampler(20)
- scored higher in this pairwise comparison
- Performance of HotKnots
- scored lower in this pairwise comparison
- Compile and download dataset for RNASampler(20) & HotKnots [.zip] - may take several seconds...
Overview
| Metric |
RNASampler(20) |
|
HotKnots |
|---|
| MCC |
0.692 |
>
|
0.548 |
| Average MCC ± 95% Confidence Intervals |
0.707
±
0.111
|
>
|
0.557
±
0.151
|
| Sensitivity |
0.621 |
>
|
0.585 |
| Positive Predictive Value |
0.776 |
>
|
0.520 |
| Total TP |
361 |
>
|
340 |
| Total TN |
68064 |
>
|
67875 |
| Total FP |
165 |
<
|
373 |
| Total FP CONTRA |
42 |
<
|
112 |
| Total FP INCONS |
62 |
<
|
202 |
| Total FP COMP |
61 |
>
|
59 |
| Total FN |
220 |
<
|
241 |
| P-value |
5.06544643719e-08 |
Performance plots
-
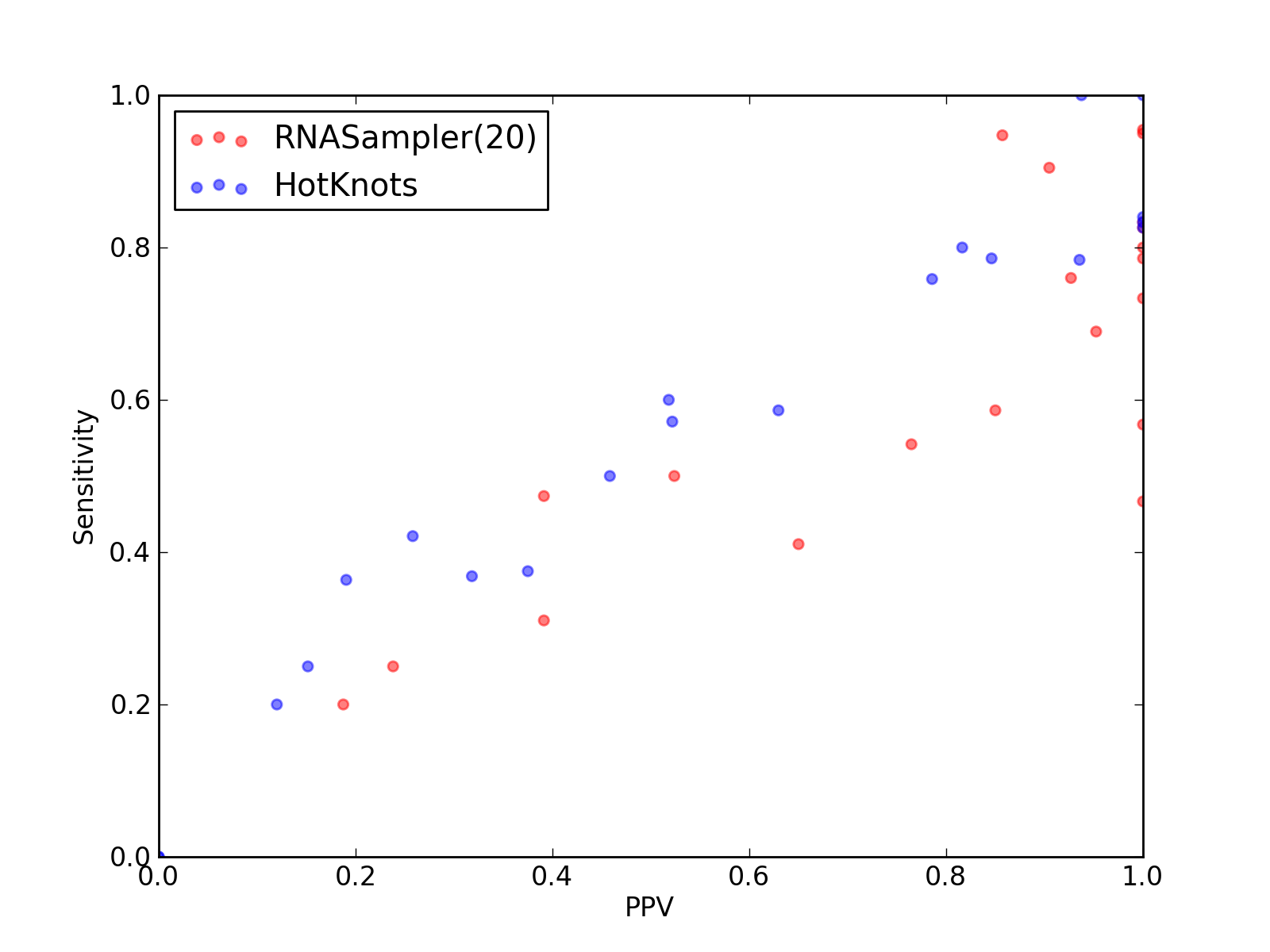
Comparison of performance of RNASampler(20) and HotKnots. Positive Predictive Value (PPV) is plotted against sensitivity. Each dot represents a single test of each method. See tables below for raw data
(individual counts for RNASampler(20)
and HotKnots).
-
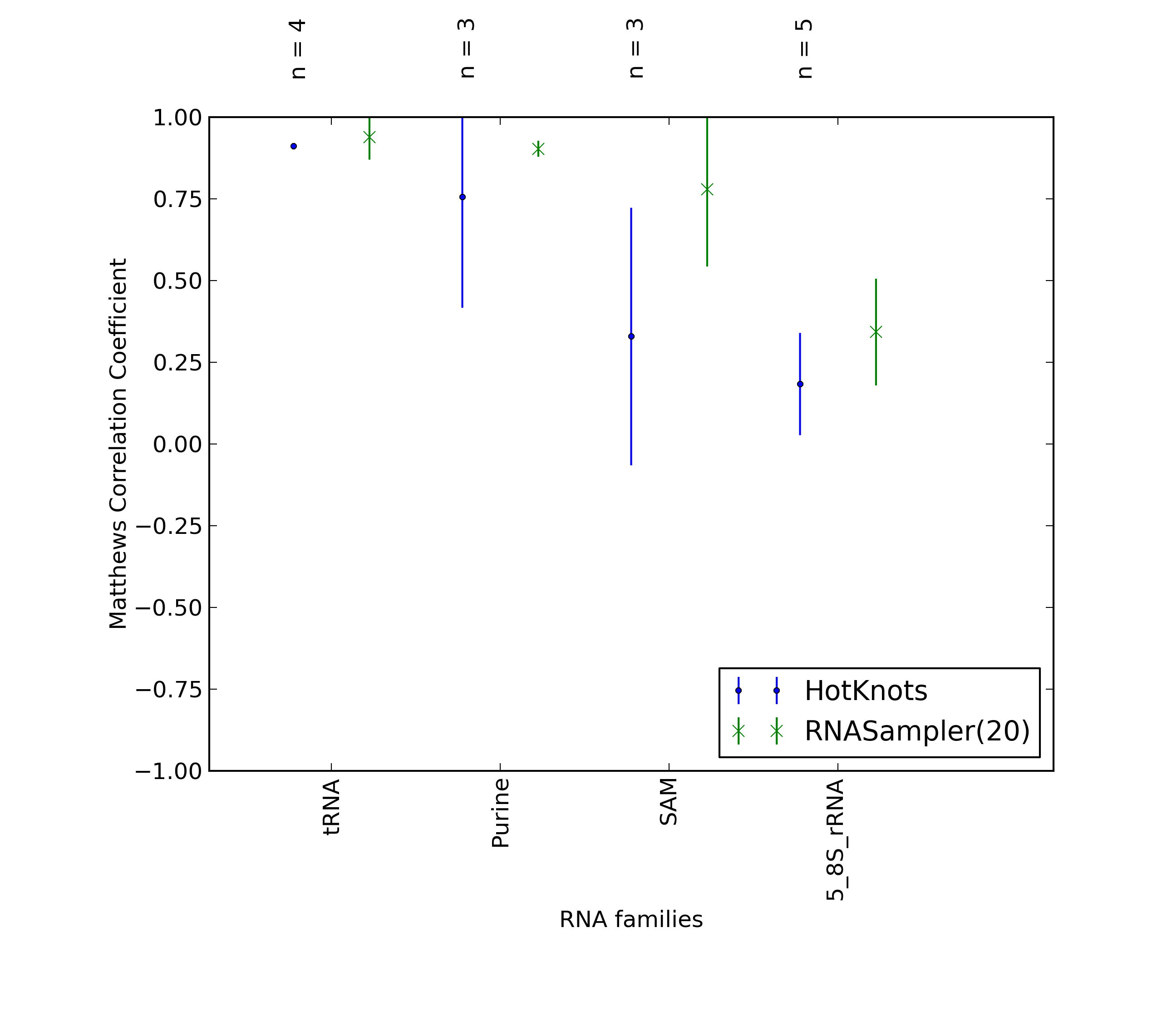
Average Matthews Correlation Coefficients (MCC) with 95% confidence intervals (CIs) were plotted for different RNA families, for which at least 3 members were present in the benchmarking dataset. 'n' denotes the number of MCCs used to calculate the average and CI. See tables below for raw data
(individual counts for RNASampler(20)
and HotKnots).
-
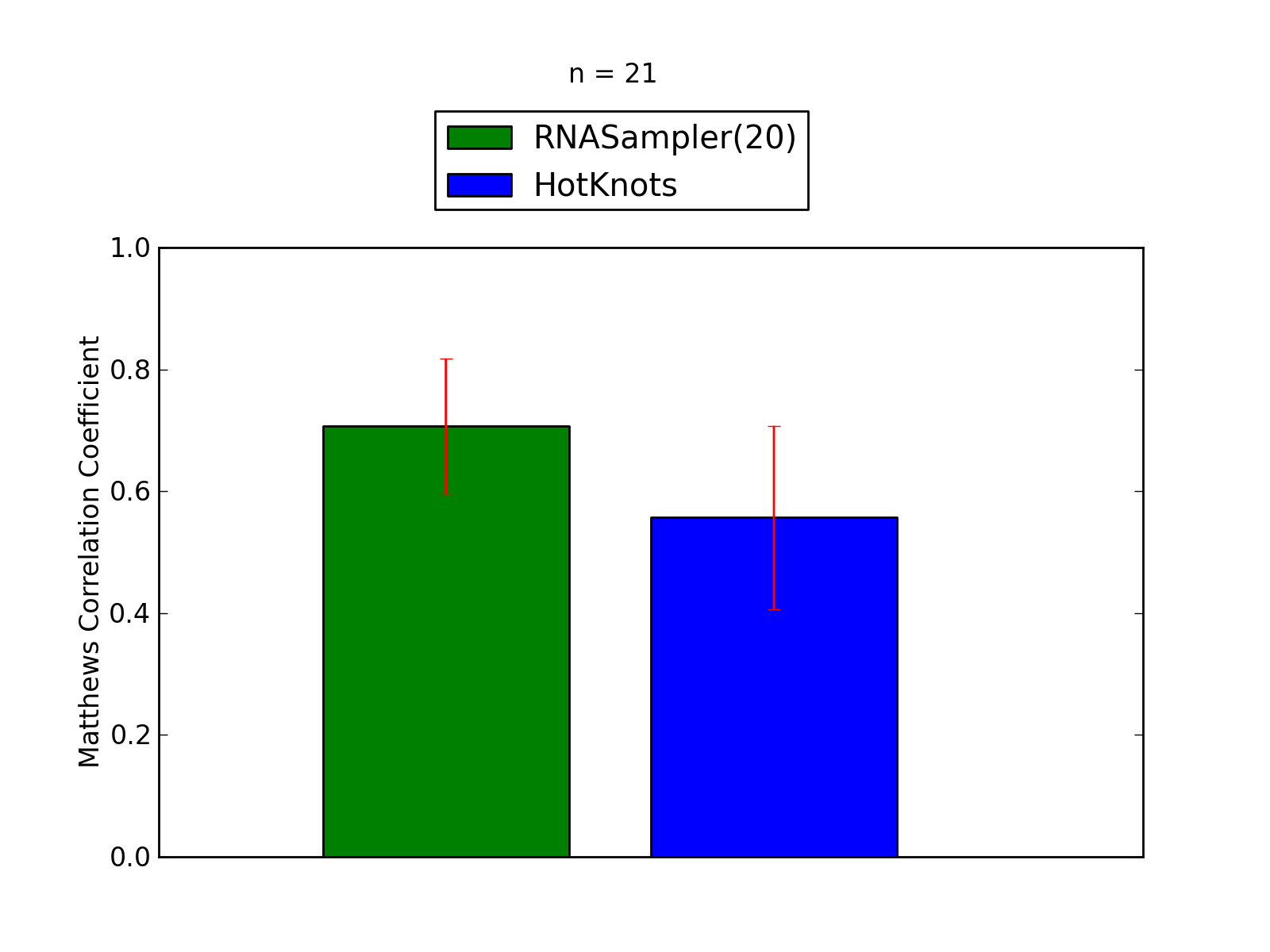
Comparison of average Matthews Correlation Coefficients (MCCs) for RNASampler(20) and HotKnots. The whiskers correspond to 95% confidence intervals (CIs). 'n' denotes the number of MCCs used to calculate average MCCs and CIs. See tables below for raw data
(individual counts for RNASampler(20)
and HotKnots).
Performance of RNASampler(20)
- scored higher in this pairwise comparison
1. Total counts & total scores for RNASampler(20)
| Total Base Pair Counts |
|---|
| Total TP |
361 |
| Total TN |
68064 |
| Total FP |
165 |
| Total FP CONTRA |
42 |
| Total FP INCONS |
62 |
| Total FP COMP |
61 |
| Total FN |
220 |
| Total Scores |
|---|
| MCC |
0.692 |
| Average MCC ± 95% Confidence Intervals |
0.707
±
0.111
|
| Sensitivity |
0.621 |
| Positive Predictive Value |
0.776 |
| Nr of predictions |
21 |
Performance of HotKnots
- scored lower in this pairwise comparison
1. Total counts & total scores for HotKnots
| Total Base Pair Counts |
|---|
| Total TP |
340 |
| Total TN |
67875 |
| Total FP |
373 |
| Total FP CONTRA |
112 |
| Total FP INCONS |
202 |
| Total FP COMP |
59 |
| Total FN |
241 |
| Total Scores |
|---|
| MCC |
0.548 |
| Average MCC ± 95% Confidence Intervals |
0.557
±
0.151
|
| Sensitivity |
0.585 |
| Positive Predictive Value |
0.520 |
| Nr of predictions |
21 |
Matthews Correlation Coeffient, Sensitivity and Positive Predictive Value have been calculated based
on the paper by Gardener & Giegerich, 2004.
| 



