

| | |||||||||||||||||
| |||||||||||||||||
Modeling example: Group I intron
The work presented here is directly based upon and would not have been possible without the efforts of the Russ Altman group:
Flores SC, Wan Y, Russell R, Altman RB. Predicting RNA structure by multiple template homology modeling. Pac Symp Biocomput. 2010:216-27. PMID: 19908374.
Problem description
A model for the group I intron RNA from Azoarcus sp. is to be built. In the PDB, a homologous structure from the Staphylococcus phage Twort, (PDB-ID 1Y0Q) is present and can be used as a template. For modeling a crucial tertiary interaction at two sites not present in the template structure, the Tetrahymena group I intron (PDB-ID 1GID) will be used as a second template to model these two regions.
The modeling of the Azoarcus intron using RNABuilder has been described in detail by Altman et al. Here, we show how the same model can be obtained with ModeRNA.
Description of the models
We have built models of the group I intron using three different alignments:
- An expert alignment created according to the instructions in the paper by Altman et al.
- An alignment obtained by the automatic procedure Infernal.
- An alignment obtained by the automatic procedure Clustal.
The models were built using four different strategies:
- Automatic model building with ModeRNA using the Twort template and an alignment.
- Refining the automatic model with ModeRNA by modeling the two tertiary interactions from a second template (see below).
- Automatic model building with RNABuilder, but using only information from the alignment (superimposed residues). RNABuilder can also include base pairing interactions, and this would surely include the models, but here we tried the alignment information only for matters of comparability.
- Using the model generated by Altman et al. using expert knowledge with RNABuilder as described in their 2010 article.
File downloads
Modeling the structural core
A first model can be built using the standard protocol of ModeRNA. What is required is the template structure [ download 1Y0Q ], and an alignment [ download FASTA ] with the target sequence. The alignment was prepared according to the literature reference above.
Because the alignment contains some large gaps, the extended fragment library is neccessary. [ see download ]. Once all files are set up, ModeRNA is able to execute the following script:
from moderna import *
a = load_alignment('group1_intron.fasta')
t = load_template('1Y0Q.pdb','A')
m = create_model(t,a)
renumber_chain(m, '1')
m.write_pdb_file('core_model.pdb')
Modeling the tertiary loop interactions
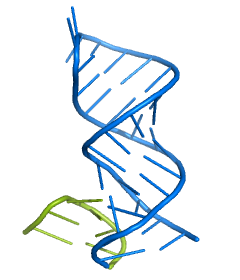
The basic protocol fails to correctly model the interaction between the loops L9/P5 and L2/P8. To include them, a template with the correct interaction was extracted from the Tetrahymena group I intron (PDB-ID 1GID) and a fragment that can be used for both sites in the Azoarcus model was extracted [ download PDB ].
In this particular example, there is a characteristic G-C base pair and a sequence motif in both the Tetrahymena fragment and the target sequence. Thus, it is not difficult to identify the residues to which the fragment needs to be fitted. To make the fit more accurate, we will superimpose two residues on each side, i.e. residues 149, 150, 153, 154, 222, 223, 250, and 251 from the extra template.
There are four ends that need to be fitted to the core structure, so we cannot use the standard fragment insertion procedure in ModeRNA (which assumes a fragment has just two ends). Also, the long stem-loop from Tetrahymena needs to be cut a little shorter, and the right sequence needs to be applied. The solution is the following script that superimposes the according residues explicitly:
from moderna import *
from moderna.ModernaSuperimposer import ModernaSuperimposer
TETRAHYMENA_LINK_SITE = ['149', '150', '153', '154', '222', '223', '250', '251']
# alignment with 14nt loop around resi 105
L2P8_SITE = ['20', '21', '24', '25', '153', '154', '170', '171']
L9P5_SITE = ['195', '196', '199', '200', '57', '58', '80', '81']
def prepare_tetrahymena_loop(start_resi, end_resi, sequence):
"""Cuts the template loop shorter and changes its sequence."""
t = load_model('1GID_excerpt.pdb')
apply_indel(t, start_resi, end_resi, Sequence(sequence))
return t
def add_tetrahymena_loop(t, m_link_site, model, seq):
"""Adds a prepared loop to the model."""
m_linker = [m[resi] for resi in m_link_site]
t_linker = [t[resi] for resi in TETRAHYMENA_LINK_SITE]
sup = ModernaSuperimposer(moved_atoms=t.get_all_atoms())
sup.get_atoms(t_linker, ["C4'", "C3'", "C1'", "N*" ], 'moved')
sup.get_atoms(m_linker, ["C4'", "C3'", "C1'", "N*" ], 'fixed')
an5, an3 = m_link_site[-3], m_link_site[-2]
frag = ModernaFragment53('residues', t['223':'250'],
anchor5=m[an5], anchor3=m[an3],
new_sequence=Sequence(seq), strict=0)
insert_fragment(m,frag)
# ---------------------------- main script --------------------------------------
m = load_model('core_model.pdb')
# model the L2/P8 loop interaction
t = prepare_tetrahymena_loop('229', '245', 'AAAC')
add_tetrahymena_loop(t, L2P8_SITE, m, 'UAAGGCAAACGCUAU')
# model the L9/P5 loop interaction
t = prepare_tetrahymena_loop('232', '242', 'GCCC')
add_tetrahymena_loop(t, L9P5_SITE, m, 'UAAGCGCCCGCCCGGGCGUAU')
fix_backbone(m)
m.write_pdb_file('model/moderna_manual_both.pdb')
|
| Copyright© - Adam Mickiewicz University - All rights reserved |