

| |
||||||||
|
||||||||

MutT enzyme (pdb: 1TUM)
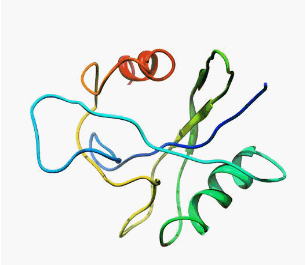
Instruction for producing the image:
Run UCSF Chimera, and then inside of the Chimera:
a) 'File'->'Fetch by ID' and in pop up type '1tum' in ID part (from PDB databese, already marked by default)
Note: 1tum is a MutT pyrophosphohydrolase-metal-nucleotide-metal complex obtained by NMR, the file contains 16 models
b) 'Select'->'Chain'->'Chain'->'A'->'1tum (#1.0)' - select first NMR model
c) 'Select'->'Invert (all models)' - invert the selection, this will select models 2-16
d) 'Actions'->'Atom/Bonds'->'delete' - remove models 2-16
e) 'Presets'->'Interactive (ribbons)' - the protein ribbon in rainbow-colored mode from blue at the N-terminus to red at the C-terminus
f) 'Actions'->'Atom/Bonds'->'hide' - hide all atoms
g) 'Presets'->'Publication 3 (depth-cued, rounded ribbon)'
h) 'File'->'Save image' - save it as a PNG file e.g. 1tum.png
Contact: Lukasz Kozlowski
If you find MetaDisorder useful, please consider citing the reference that describes this work:
Kozlowski LP, Bujnicki JM. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012 May 24;13(1):111.
This work was supported by Polish Ministry of Science and Higher Education (grant NN301 190139).