

| |
||||||||
|
||||||||

Overview
GeneSilico MetaDisorder web service provides access to four metapredictors designed to predict disorder in proteins using existing bioinformatics tools and machine learning. As an output you will get metapredictions based on optimized results from primary methods. Methods were carefully benchmarked and validated using our own dataset and during blind tests CASP8 and CASP9.MetaDisorder
This method uses 15 predictions from primary disorder predictors (for a complete list see table below) and than weights their output according method accuracy (see Figure 1). Individual weights of methods correspond to 1+Sw score of the method for the first dataset. Although this method seems to be very simple, it has been proven during CASP (the best method in 2008) that this approach can be very succesful (AUC for targets in CASP8 was over 0.91!!!). On the other hand, those targets were very easy to predict (AUC for our own, more challenging dataset was ~0.83, which seems to be more realistic).Metadisorder3D
Uses alignments from 8 fold recognition method (for a complete list see table below). The idea behind it is that the gaps in alignments can be used as an indicator of disorder (but please compare easy case in Example 1 with more blurred one in Example2). Resulting predictor was able to detect disorder with AUC 0.83 compared with 0.91 for CASP8 targets and MetaDisorder). It must be stressed that each method can have different utility for disorder prediction and additionally alignments produced by FR methods are scored according their accuracy (this mean that the information from "good" alignments should be scored more than from worse). To address those questions we optimized the weights for given method taking into account alignment quality using genetic algorithm implemented in PyEvolve. CASP8 target were used for training and benchmarking. Additionally, the method uses smoothing filter (to avoid one or two letter long miss predictions) and the correction for terminal amino acids (prone to be more disordered).MetaDisorder3D takes at most 10 best alignments produced by FR method and divide them to "good", "medium" and "poor" alignments using methods native scores:
MetadisorderMD
Merges above mentioned methods into one using genetic algorithm. Trained on CASP8 targets, validated during CASP9MetadisorderMD2
Similar to previous method, but for the optimization, instead of using Sw score in fitness function of GA, we use so-called Sww score, which tries to capture the best features of AUC and Sw score. Sww score is calculated exactly as classical Sw, but the disorder/order thresholds are moved like it is done in AUC.Read more:


Dataset used to train and benchmark methods
We used 3 separate datasets which were cross validated and/or bootstrapped when used.1) 1147 proteins from PDB database filtered by resolution <2 Å, R-factor < 0.2, length 50-1000 amino acids and sequence identity < 20%
2) 566 proteins composed from Disprot (version 3.6) and CASP7 targets
3) 122 targets from CASP8
How disorder is defined and calculated?
Prediction DDDD----------------------------------DDDDDDD---DDD
TP - both disorder DDD
FN - predicted as ordered, but is disordered D-D
TN - both ordered ---
FP - predicted as disordered, but is ordered -D-
The accuracy of methods was calculated using Sw score and AUC (area under curve):
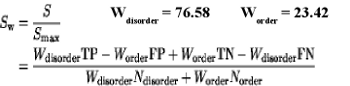
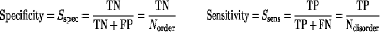
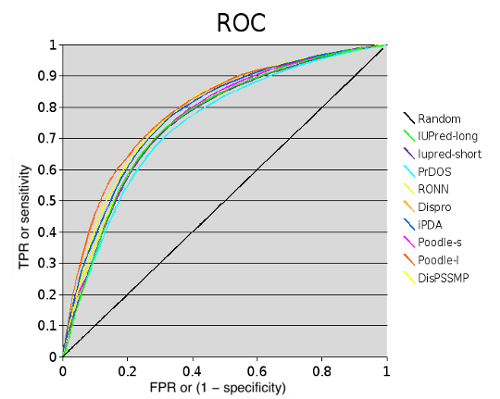
| Method | Sw | ROC |
|---|---|---|
| poodle-l | 0.453 | 0.792 |
| ipda | 0.439 | 0.779 |
| iupred-l | 0.417 | 0.759 |
| dispro | 0.415 | 0.761 |
| poodle-s | 0.414 | 0.763 |
| iupred-s | 0.412 | 0.757 |
| Spritz-l | 0.399 | - |
| prdos | 0.391 | 0.745 |
| ronn | 0.380 | - |
| DISOPRED2 | 0.367 | - |
| DisEMBL | 0.328 | - |
| Spritz-s | 0.312 | - |
| MetaDisorder (binary) | 0.464 | 0.801 |
| MetaDisorder (continous) | 0.469 | 0.821 |
Primary methods for disorder prediction
Fold recognition methods for disorder prediction
This site is powered by:



