

| | |||||||||||||||||
| |||||||||||||||||
Python Architecture
Updated and rewritten for ModeRNA 1.4 (March 2010)
Disclaimer: This page is written for programmers. Expect lots of tech lingo.
All commands in ModeRNA are technically pure Python functions. This allows ModeRNA to cooperate with any Python code. For normal scripting it is enough to import the moderna module to have all commands available. For more sophisticated operations, we give a brief introduction to the program architecture here.
Test suite
In the test/ directory of the source distribution, acceptance and unit tests using unittest are deposited. To run the test suite, type:
python test_ModeRNA_all.py
In the same way, a particular test set can be checked (e.g. for the Alignment class):
python test_alignment.py
This is particularly useful if you are trying to customize the program and want to know whether you disabled any of its functionalities
Overview of the class structure
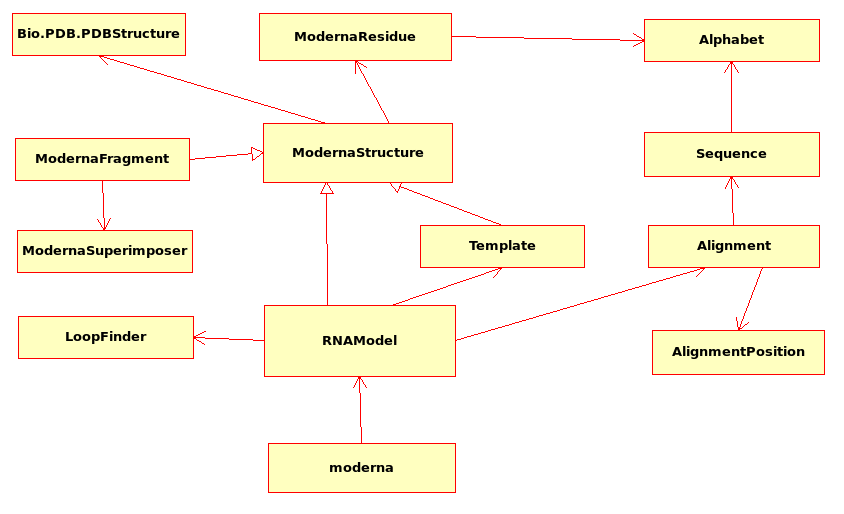
UML class diagram: The diagram represents each class as a box. Normal arrows refer to dependencies, triangle arrows to inheritance (from the child to the parent class). Only the most important classes are represented here.
Module moderna.moderna
Implements the Facade Pattern. For each documented command, there is a convenience function here. This is the only part of ModeRNA, where we guarantee API stability.
Class moderna.ModernaAlphabet.Alphabet
Instances of this class are a dict with the nomenclature scheme for modified and unmodified nucleotides. Keys are the long abbreviations, e.g. A, m1A, m66Am, values are
AlphabetEntry objects containing short abbreviation, pdb abbreviation, full name, and original base.
The data used by this class is in moderna/data/modifications_names_table.
Class moderna.ModernaAlignment.Alignment
Reads an alignment in fasta format from a string or file and creates a list of AlignmentPosition objects out of it.
All positions are automatically categorized into lists determining what will happen to them during modeling:
- copy: template and target are identical and not ANY_RESIDUE (dot).
- copy_backbone: target or template contain the ANY_RESIDUE (dot) symbol.
- exchange: target and template differ but are not modified.
- remove_modifications: template is a modified nucleotide but target is not.
- add_modification: target is a modified nucleotide but template is not.
- add_fragment: list of positions belonging to an indel, including flanking residues.
- add_fragment_5p: indel positions at the 5’ end.
- add_fragment_3p: indel positions at the 3’ end.
- difficult: ModeRNA doesn’t know what to do at a particular position.
Positions in the alignment can be addressed by:
- Alignment[index] : index refering to the entire alignment (including gaps).
- Alignment.target_numeration[index] : index refering to position in the target sequence.
- Alignment.template_numeration[index] : index refering to position in the template sequence.
Class moderna.ModernaResidue.ModernaResidue
Implements the Decorator Pattern on Bio.PDB.Residue objects.
It supplements functions to:
- recognize modified nucleotides
- exchange bases
- add modifications
- remove modifications
- recognize backbone breaks
- recognize backbone atoms clashing with other atoms
- rotate the chi angle
Class moderna.ModernaStructure.ModernaStructure
This class wraps a Bio.PDB.Chain object. Despite the name it is responsible for one RNA chain only but this is likely to change. It can be iterated, and returns ModernaResidue objects. Residues can also be accessed per m[x] by their numbers.
Class moderna.Template.Template
This is a subclass of ModernaStructure for template structures. Its main purpose is to map the indices from the template sequence in Alignment to residues in the template structure.
Class moderna.RNAModel.RnaModel
This subclass of ModernaStructure does all the modeling work, especially in the automatic mode. It needs a Template and Alignment object to carry out its job. The most important method is probably create_model(), the second most important insert_fragment().
Module moderna.ModernaFragment
ModernaFragment classes handle small pieces of structure that can be added to an RNAModel. They contain mechanics to superimpose a fragment, remodel the residues at its ends (referred to flanking, stem, or anchor residues), exchange the sequence of the fragment, renumber residues, and fix eventual backbone breaks.
The following classes are available:
ModernaFragment:Implements the Template Method pattern. It is an abstract superclass for all fragments.ModernaFragment5:fragment with an anchor residue at its 5— end.ModernaFragment3:fragment with an anchor residue at its 3— end.ModernaFragment53:fragment with an anchor residue at both ends (the one used for indels in the middle of a structure.
Class moderna.SearchLir.FragmentFinder
Implements the Builder Pattern to find aproppriate fragments from the fragment library.
Class moderna.ModernaSuperimposer.ModernaSuperimposer
Wrapper for Bio.PDB.Superimposer, that can
extract atoms with predefined names from residues.
Allows to rotate and translate atoms in space. Used to fit fragments during remodeling single residues and fragments.
|
| Copyright© - Adam Mickiewicz University - All rights reserved |