

| |
|
|||||||
|
||||||||
| |
||||||||
The tale of protein disorder
Even today, when people think about the protein structures, in most cases they sees them like that:
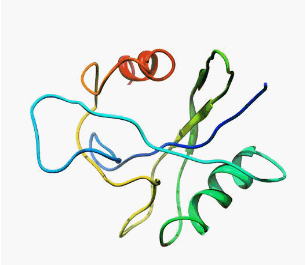 |  |
| MutT enzyme (pdb: 1TUM) | Human
pancreatic ribonuclease (pdb: 2k11) |
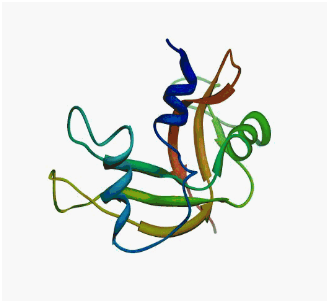
 |
 |
| Human
tRNA endonuclease SEN15 subunit (pdb: 2gw6) | Kalirin
DH1 domain (pdb: 1kr9) |
borring, borring, ... well defined and static structures.
But,
actually it is not true. Proteins are flexible, they can change the
shape:
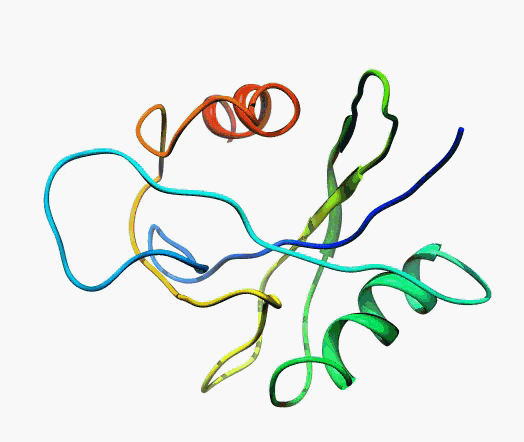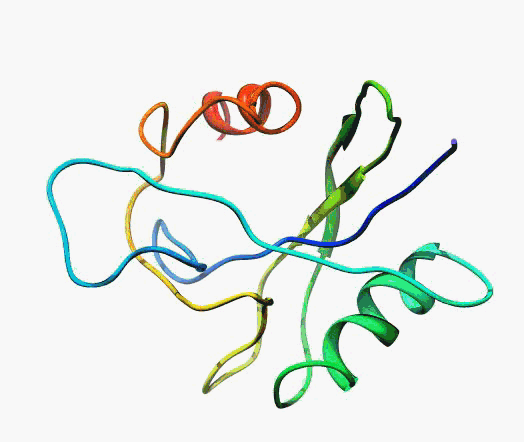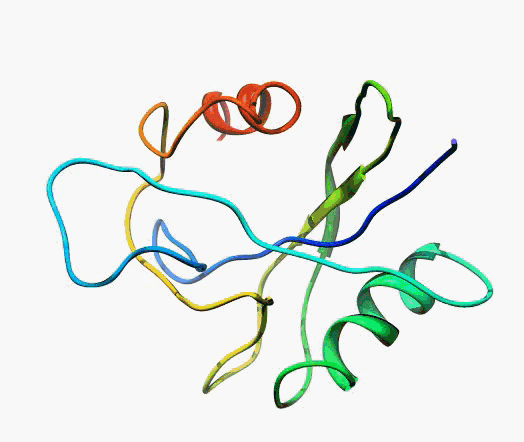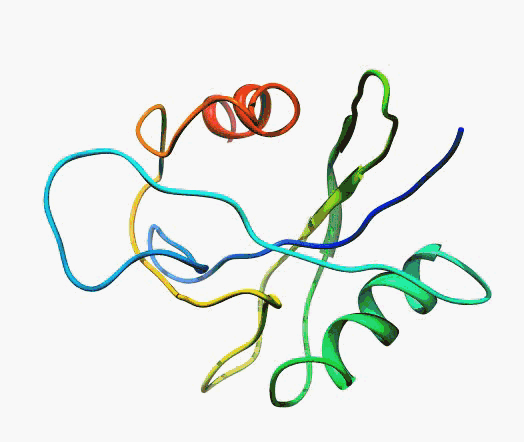 |
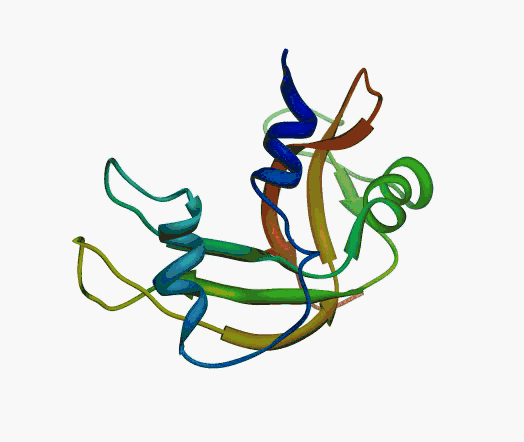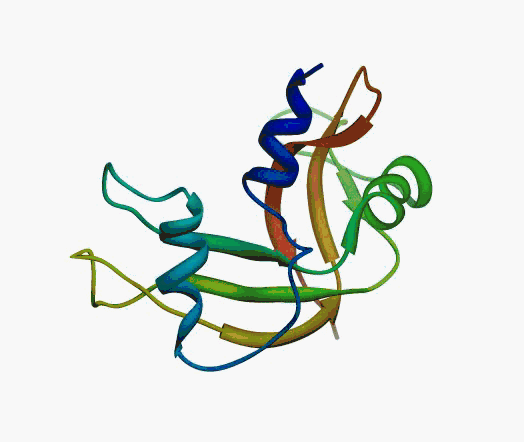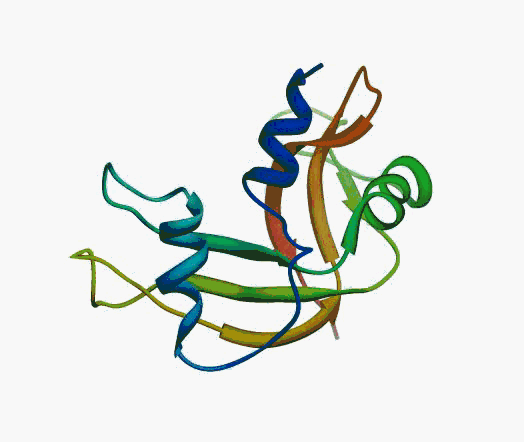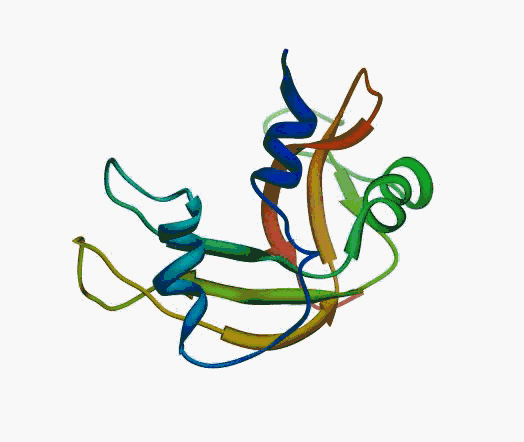 |
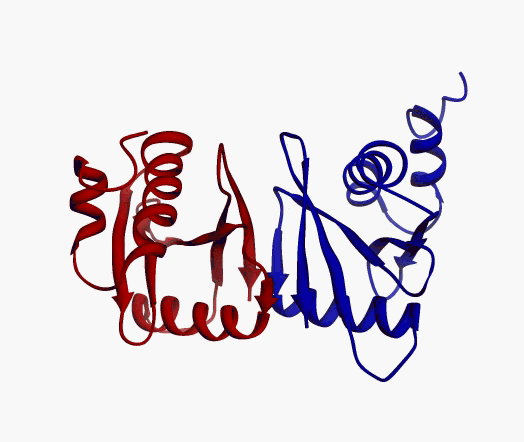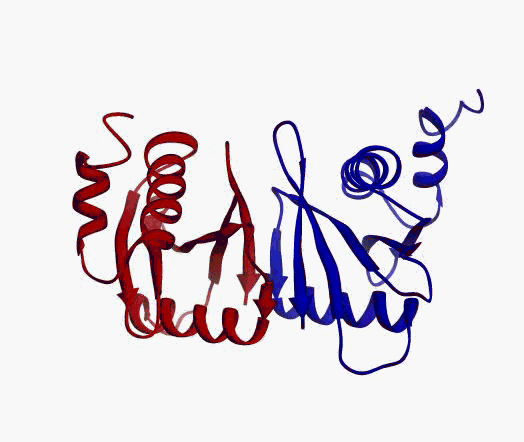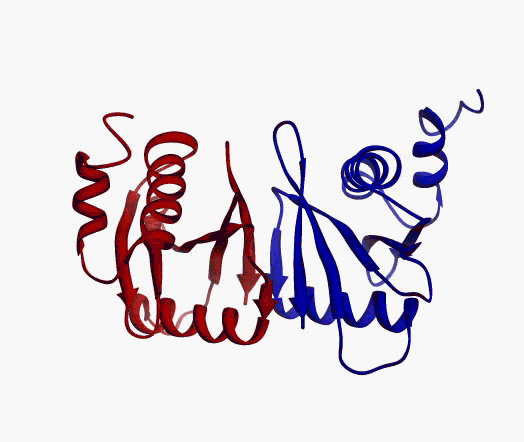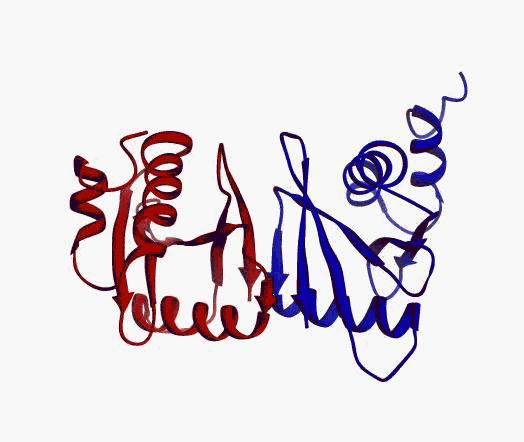 |
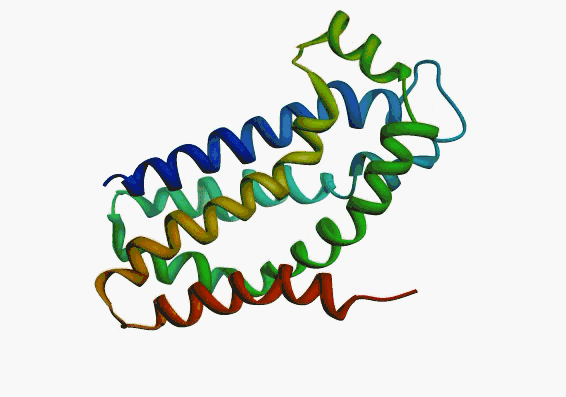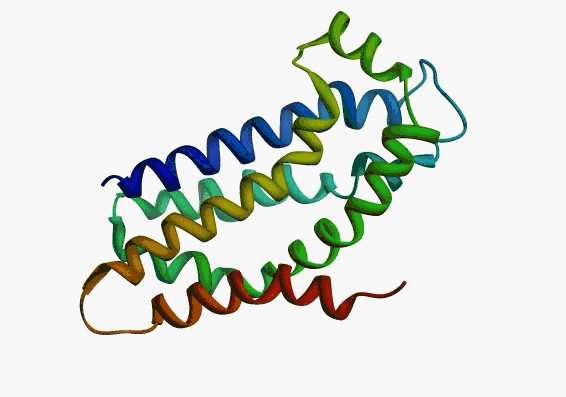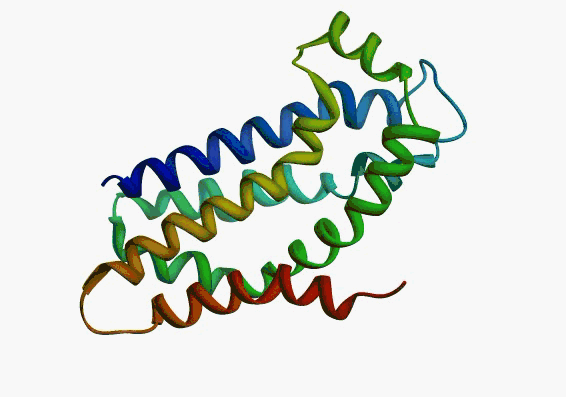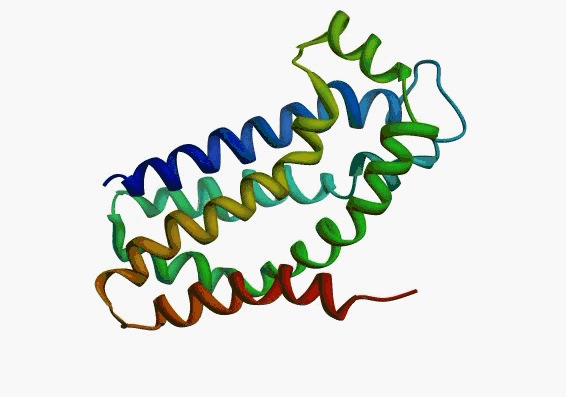 |
Even better, some of them can be extremelly flexible
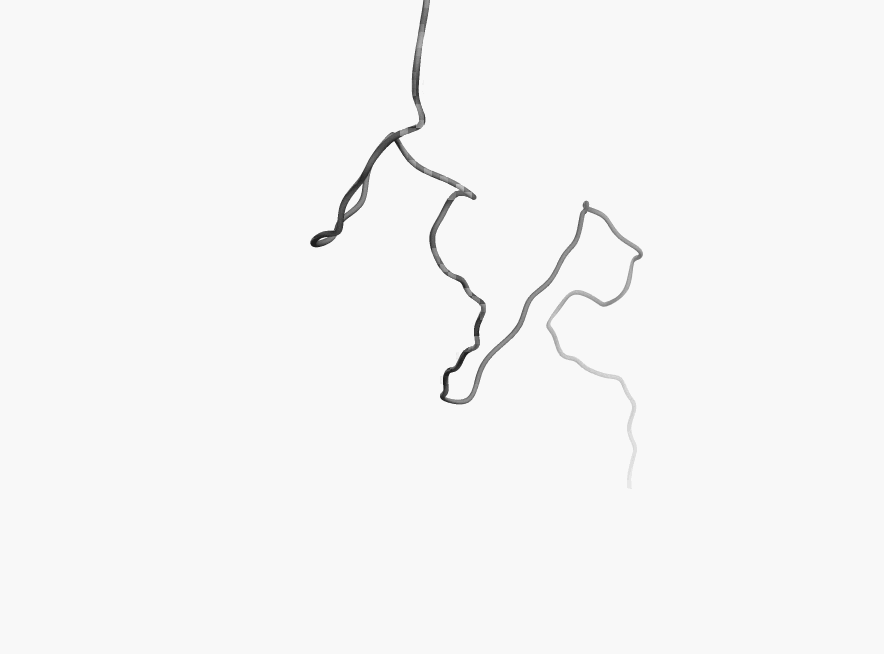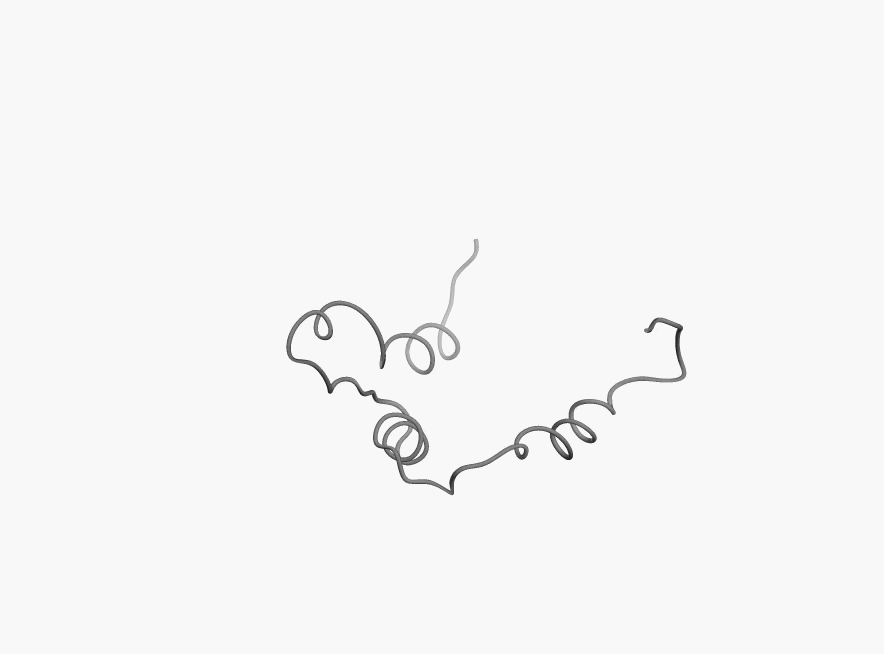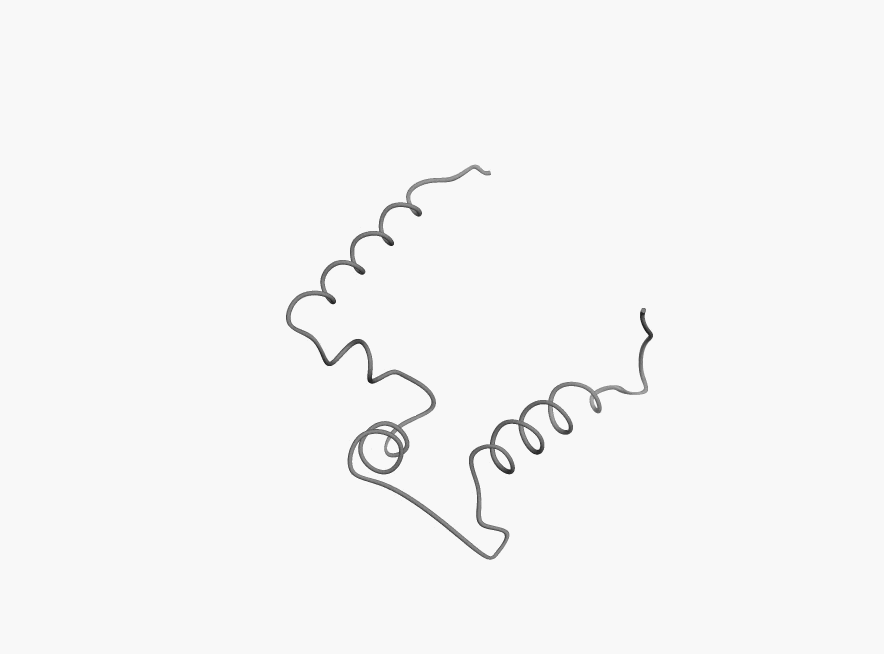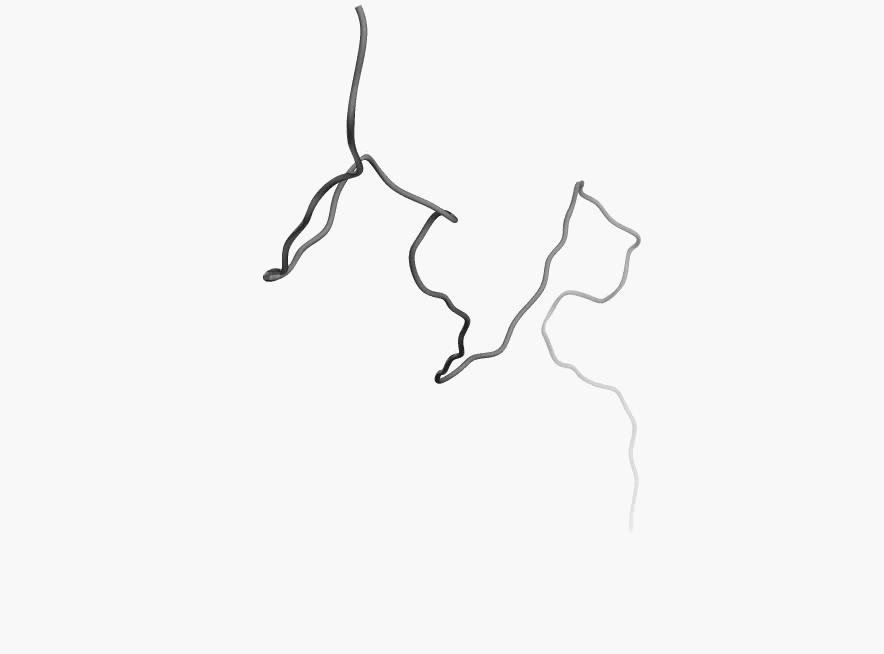
Protein
disorder at the NTD of the target T0590 (PDB id: 2kzw)
We call them intrinsically
unstructured proteins (IUPs)
or simply unfolded proteins or disordered proteins. The
discovering of IUPs chalanged one of the main protein structure
paradigm, which states that a specific well-defined structure is
required for the correct function of a protein and that the structure
defines the function of the protein. This is clearly not true for
intrinsically unfolded proteins that remain functional despite the lack
of a well-defined structure. Such proteins, in some cases, can adopt a
fixed three dimensional structure after binding to other
macromolecules. Due this intrinsic flexibility protein disorder
is particularly enriched in proteins implicated in cell signaling,
transcription and chromatin remodeling functions. Moreover, they are
connected with many diseases (a lot of key oncogens have large
unstructured regions, e.g. p53 and BRCA1, D2D2 concept).
Bellow you can few more examples of disordered proteins:
Bellow you can few more examples of disordered proteins:
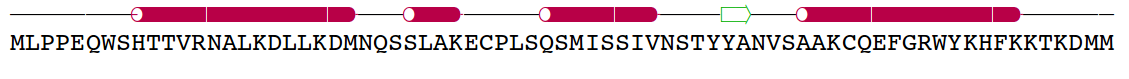
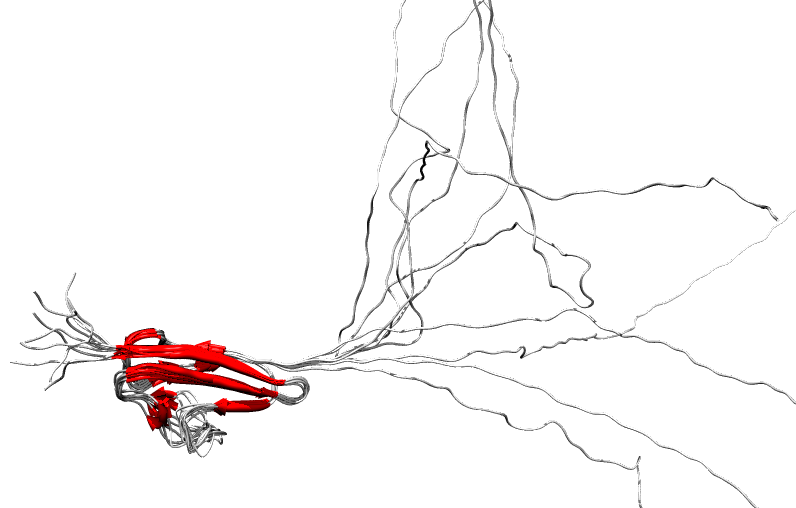
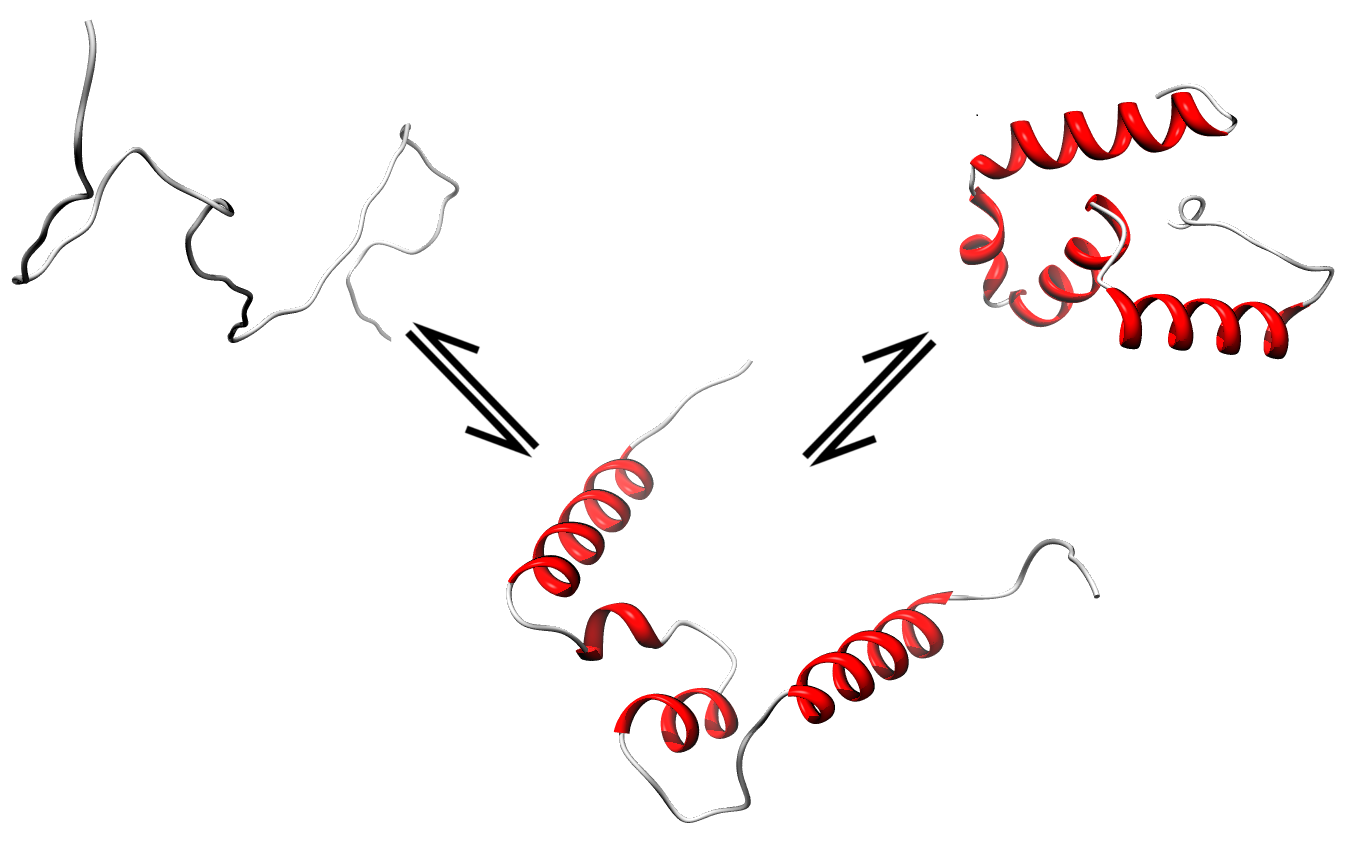 |
 |
Simulation
of folding and unfolding (transition form order/disorder state for
CASP8 target T0556 done using MODELLER based on predicted secondary
structure and remote template (PDB id: 3dnv_B).
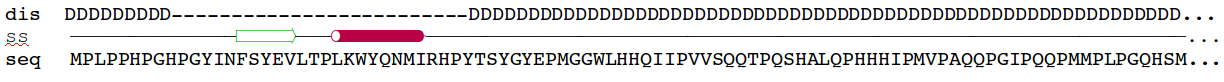
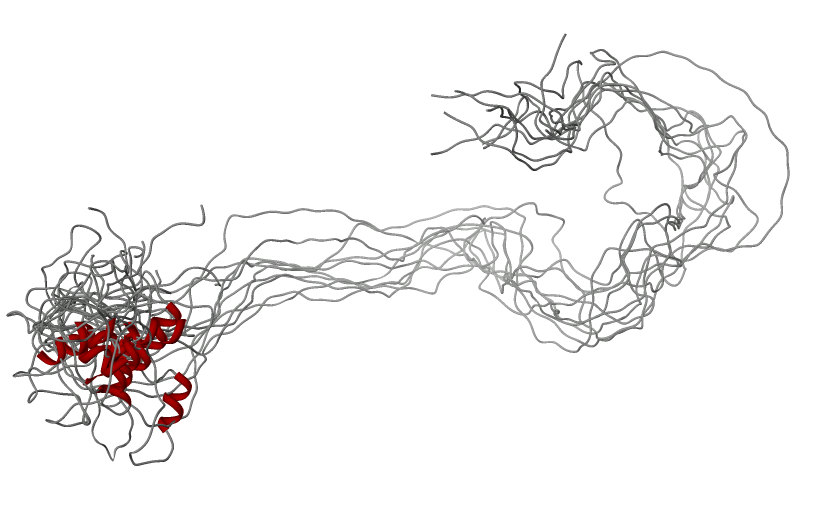
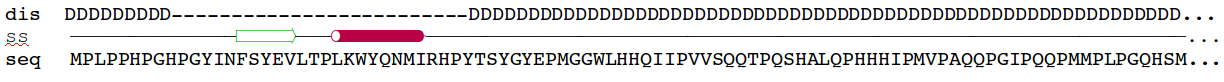

Amelogenin as an
example of intrinsically disordered
protein. Extended, unfolded state of the protein is crucial for
formation of protein-mineral interactions (generated by MODELLER with
restrains for predicted secondary structure). Above the model predicted
alignment with secondary structure is presented.
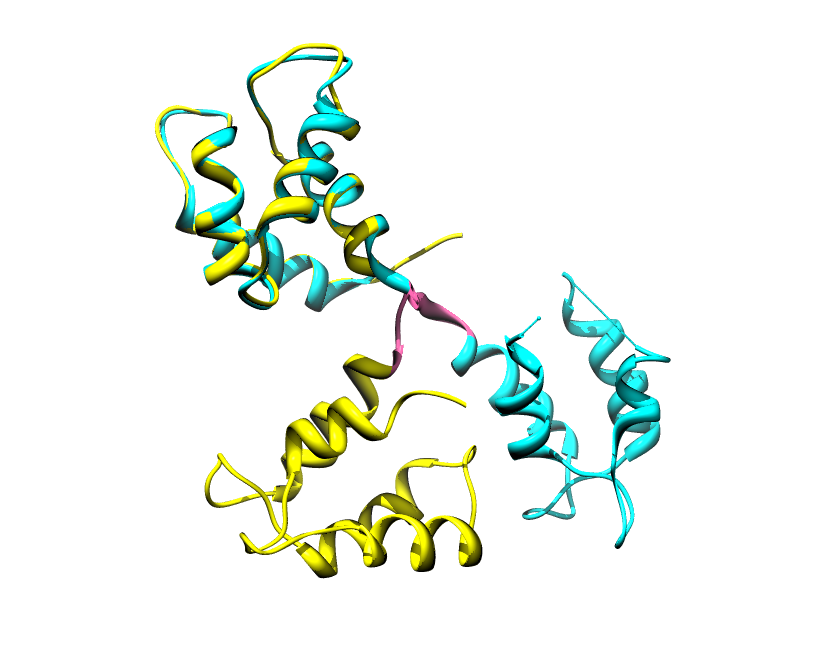
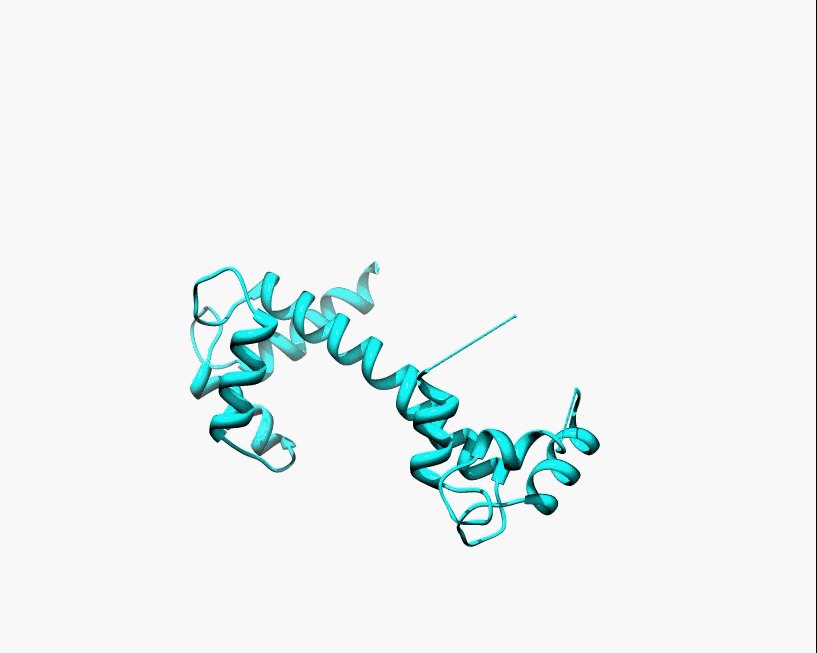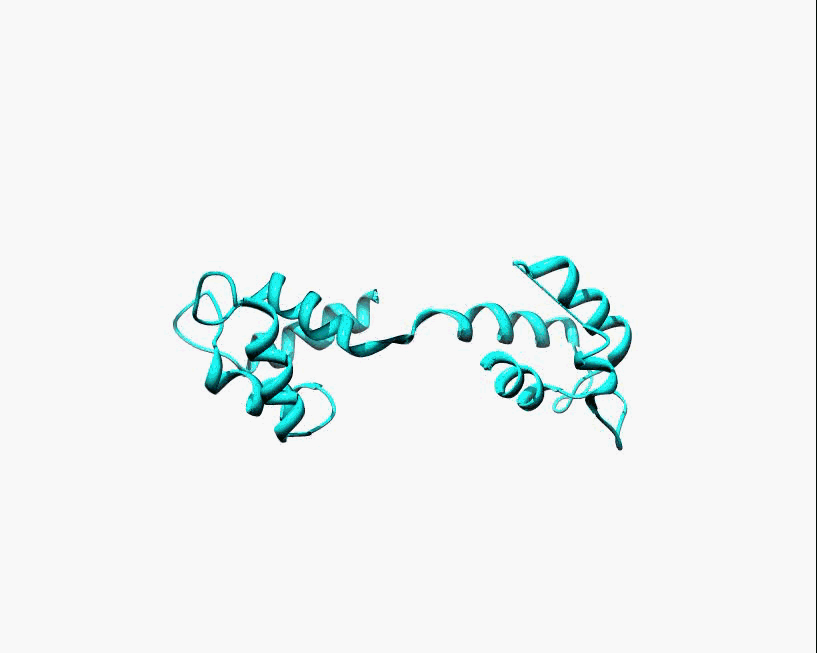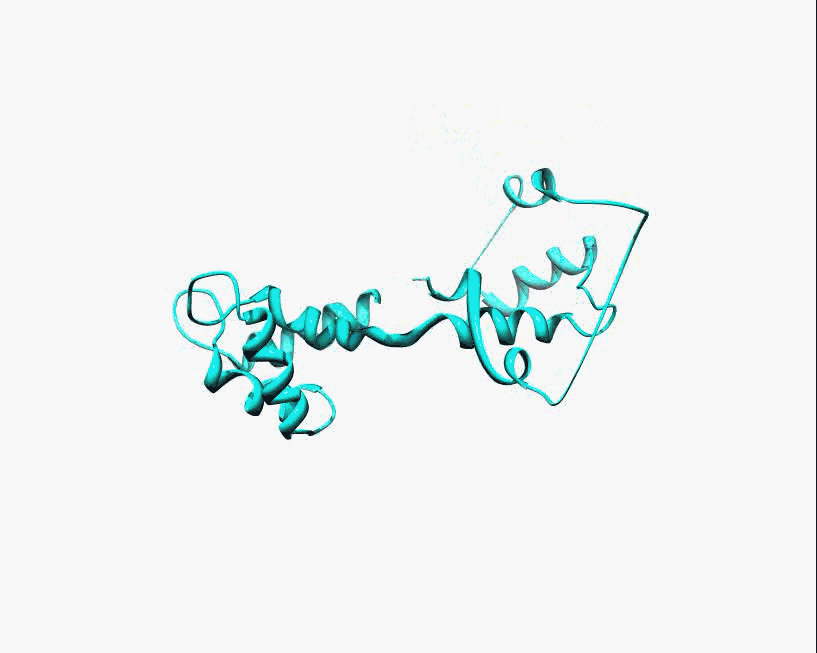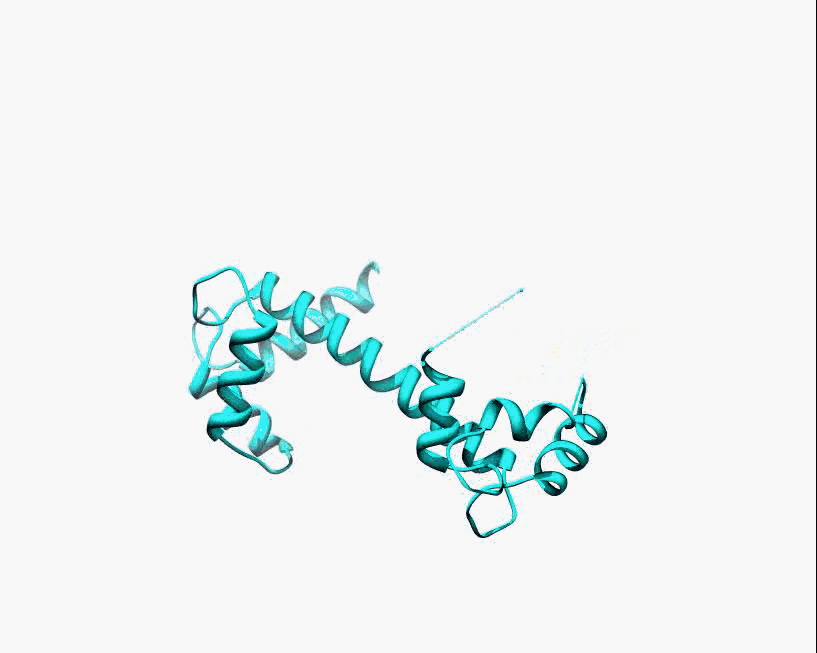
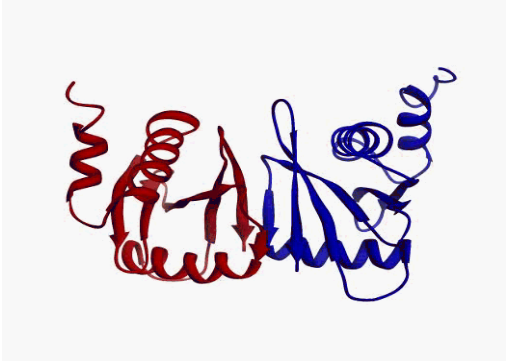
Disorder as a flexible linker. Calmodulin with and without Ca2+ ions (based on pdb: 2f2o, 1cfd, 1cll).
Contact: Lukasz Kozlowski
If you find this site useful, please consider citing the reference:
Kozlowski LP, Bujnicki JM. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012 May 24;13(1):111.
This work was supported by Polish Ministry of Science and Higher Education (grant NN301 190139).

If you find this site useful, please consider citing the reference:
Kozlowski LP, Bujnicki JM. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012 May 24;13(1):111.
This work was supported by Polish Ministry of Science and Higher Education (grant NN301 190139).
My other projects:
- IPC - protein Isoelectric Point Calculator - isoelectric point and molecular weight from protein sequence
- GeneSilico fold recognition server - development and maintenance (over 100 bioinformatics tools integrated, 3000 registered users)
- CompaRNA - continuous benchmarking of RNA structure prediction methods
- GDFuzz3D - protein contact map to 3D structure retrieval service
- Shannon entropy calculator - Real example how to calculate and interpret information entropy
- RNA metaserver - Meta-tool for prediction of RNA secondary structure
