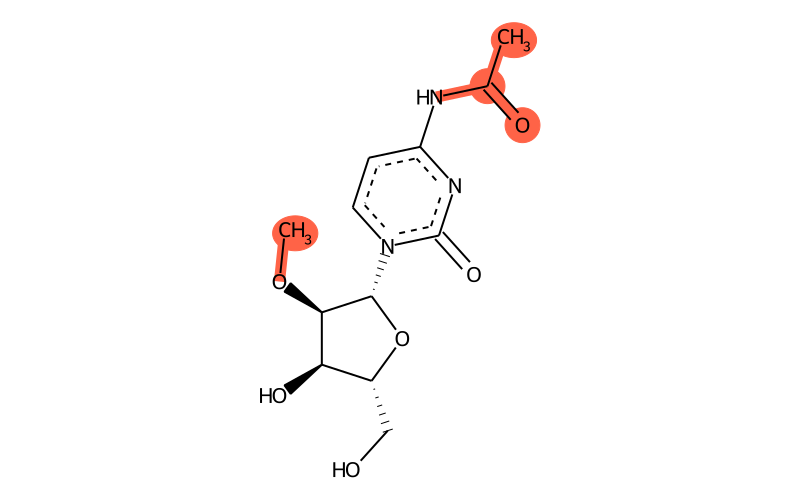
Summary
| Full name | N4-acetyl-2'-O-methylcytidine |
| IUPAC name | N-[1-[(2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxyoxolan-2-yl]-2-oxopyrimidin-4-yl]acetamide |
| Short name | ac4Cm |
| MODOMICS code new | 2000009042C |
| MODOMICS code | 042C |
| Synonyms |
113886-71-8
Ac-2-OMe-C Ac-2'-OMe-C Ac-2/'-OMe-C AC-32177 AKOS015899633 BS-19249 CID 10902616 CID10902616 CTK4A8501 CYDFBLGNJUNSCC-QCNRFFRDSA-N Cytidine, N-acetyl-2'-O-methyl- Cytidine,N-acetyl-2'-O-methyl- DTXSID00447815 HG1334 I14-11183 I14-32546 J-700206 K-1012 MFCD15145149 N-[1-[(2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxyoxolan-2-yl]-2-oxopyrimidin-4-yl]acetamide N-(1-((2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxytetrahydrofuran-2-yl)-2-oxo-1,2-dihydropyrimidin-4-yl)acetamide N4 -acetyl-2'-O-methyl cytidine N4-Acetyl-2'-O-methyl cytidine N4-Acetyl-2'-O-methylcytidine N4-ACETYL-2'-O-METHYL-CYTIDINE N-Acetyl-2'-O-methylcytidine SCHEMBL3499005 ST24045894 ZINC34556412 |
| Nature of the modified residue | Natural |
| RNAMods code | ℵ |
| Residue unique ID | 106 |
| Found in RNA | Yes |
| Related nucleotides | 257 |
Chemical information
| Sum formula | C12H17N3O6 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| SMILES | CO[C@H]1[C@H]([n]2c(=O)nc(NC(=O)C)cc2)O[C@H](CO)[C@H]1O |
| logP | -1.4597 |
| TPSA | 122.91 |
| Number of atoms | 21 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| PDB no exac match , link to the most similar ligand | LHH |
| HMDB (Human Metabolome Database) no exact match, link to the most similar ligand | None |
| InChI | InChI=1S/C12H17N3O6/c1-6(17)13-8-3-4-15(12(19)14-8)11-10(20-2)9(18)7(5-16)21-11/h3-4,7,9-11,16,18H,5H2,1-2H3,(H,13,14,17,19)/t7-,9-,10-,11-/m1/s1 |
| InChIKey | CYDFBLGNJUNSCC-QCNRFFRDSA-N |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
15951156
23146523 74998049 98580306 99375003 127694758 135368010 152039052 162776819 164216540 185986197 222062119 225039549 229509671 249865113 252225507 260419907 275348827 277383362 300407609 307087076 310277405 312343848 314988887 315978123 316541204 342465011 347266369 349560360 355189867 364167553 374045991 375228471 375396316 377643449 383452028 385027262 403995368 404623341 404823683 426483275 434737445 438515873 439484831 440703661 441133467 442078809 443532598 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
COC1C(n2c(=O)[nH]c(=NC(=O)C)cc2)OC(CO)C1O tautomer #0
COC1C(n2c(=O)nc(NC(=O)C)cc2)OC(CO)C1O tautomer #1 COC1C(n2c(=O)[nH]c(=NC(O)=C)cc2)OC(CO)C1O tautomer #2 COC1C(n2c(=O)nc(NC(O)=C)cc2)OC(CO)C1O tautomer #3 COC1C(n2c(O)nc(=NC(=O)C)cc2)OC(CO)C1O tautomer #4 COC1C(n2c(=O)nc(N=C(O)C)cc2)OC(CO)C1O tautomer #5 COC1C(n2c(O)nc(=NC(O)=C)cc2)OC(CO)C1O tautomer #6 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
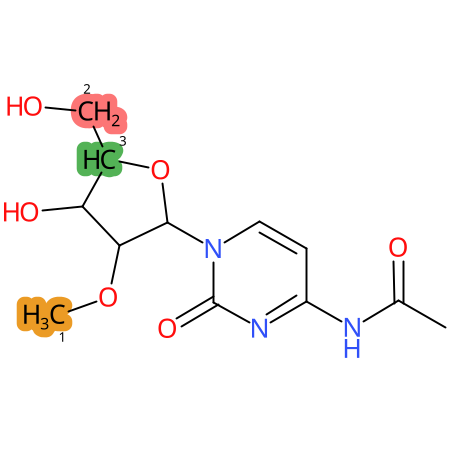
|
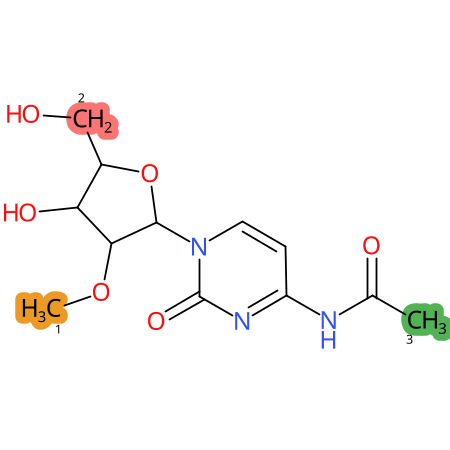
|
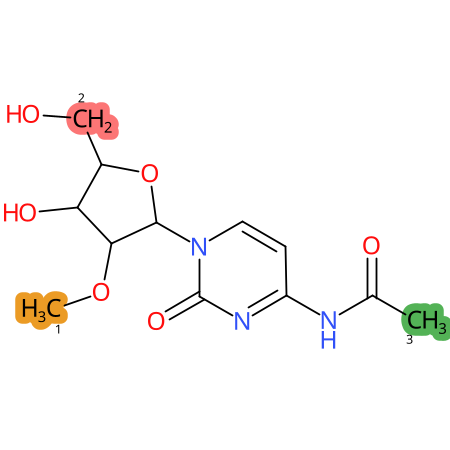
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 299.1117 |
| Average mass | 299.28 |
| [M+H]+ | 300.1195 |
| Product ions | 154/112 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at O2 |
| other | acetylamide |
Reactions producing N4-acetyl-2'-O-methylcytidine
| Name |
|---|
| Cm:ac4Cm |
| ac4C:ac4Cm |
Last modification of this entry: Sept. 22, 2023