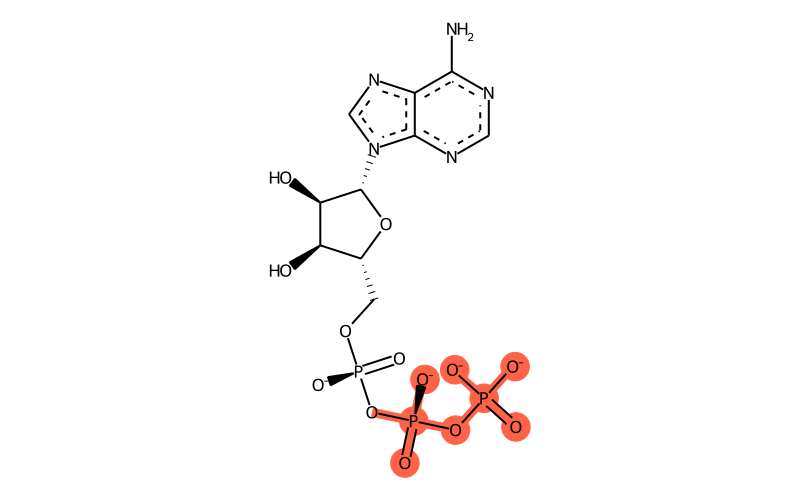
Summary
| Full name | adenosine-5'-triphosphate |
| IUPAC name | [[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-oxidophosphoryl]oxy-oxidophosphoryl] phosphate |
| Short name | pppA |
| MODOMICS code new | 2000000553A |
| MODOMICS code | 553A |
| Synonyms |
ATP(4-)
|
| Nature of the modified residue | Natural |
| Residue unique ID | 233 |
| Found in RNA | Yes |
| Related nucleosides | 109 |
| RCSB ligands |
Chemical information
| Sum formula | C10H12N5O13P3 |
| Type of moiety | nucleotide |
| Degeneracy | not aplicable |
| SMILES | [O-][P@](OP(=O)([O-])[O-])(O[P@]([O-])(OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)[n]1c[n]c2c1[n]c[n]c2N)=O)=O |
| logP | 0.705 |
| TPSA | 319.88 |
| Number of atoms | 31 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 13 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 18 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| InChI | InChI=1S/C10H16N5O13P3/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(26-10)1-25-30(21,22)28-31(23,24)27-29(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H,23,24)(H2,11,12,13)(H2,18,19,20)/p-4/t4-,6-,7-,10-/m1/s1 |
| InChIKey | ZKHQWZAMYRWXGA-KQYNXXCUSA-J |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
2157
8148096 11688362 24877532 39443564 49737792 50247497 53812597 53812679 53812681 53812739 53812902 53812904 56412694 56436522 84970651 85145927 87557452 87557505 87557712 104075349 114001216 131319136 164151282 170481218 170481265 170481338 170481406 170481407 170481627 170481825 170482177 170482179 170482197 170482263 170483168 170483402 170483906 170483907 170484256 170485046 170485047 170485048 223501249 317306102 319304259 341825608 342583024 349975437 355358160 376955710 385662821 386285271 386499327 419578700 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 78.884776196 |
| Energy [Eh]: | -2663.9583249911 |
| HOMO [eV]: | -8.4657 |
| LUMO [eV]: | 1.2451 |
| Gap [eV]: | 9.7108 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
[O-]P(OP(=O)([O-])[O-])(OP([O-])(OCC1OC(C(O)C1O)n2cnc3c2ncnc3N)=O)=O tautomer #0
[O-]P(OP(=O)([O-])[O-])(OP([O-])(OCC1OC(C(O)C1O)n2cnc3c2ncnc3N)=O)=O tautomer #1 [O-]P(OP(=O)([O-])[O-])(OP([O-])(OCC1OC(C(O)C1O)n2cnc3c2[nH]cnc3=N)=O)=O tautomer #2 [O-]P(OP(=O)([O-])[O-])(OP([O-])(OCC1OC(C(O)C1O)n2cnc3c2nc[nH]c3=N)=O)=O tautomer #3 [O-]P(OP(=O)([O-])[O-])(OP([O-])(OCC1OC(C(O)C1O)N2C=NC3C2=NC=NC3=N)=O)=O tautomer #4 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
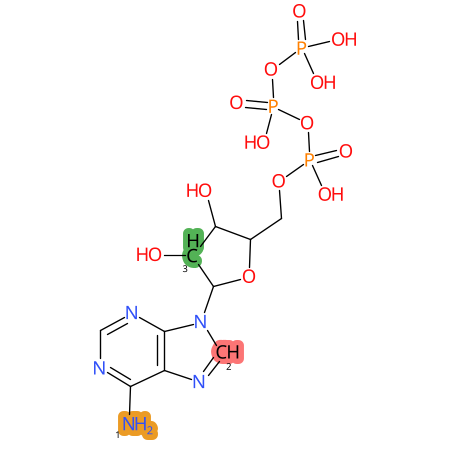
|
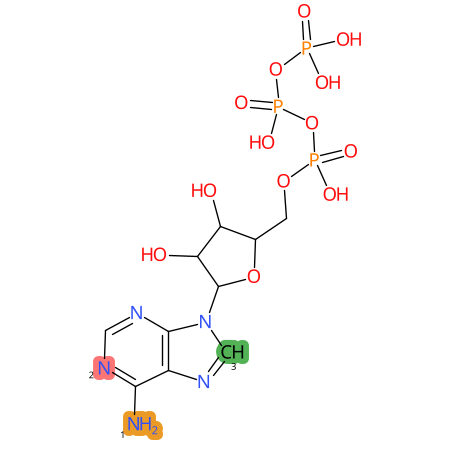
|
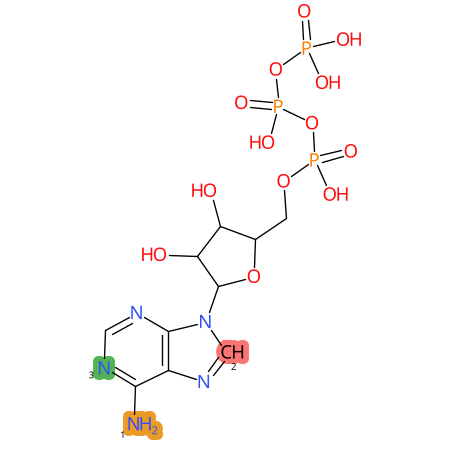
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | None |
| Average mass | 503.149 |
| [M+H]+ | not available |
| Product ions | not available |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Last modification of this entry: Sept. 15, 2025