⬤ Hydrogen (H): white
⬤ Carbon (C): gray
⬤ Oxygen (O): red
⬤ Phosphorus (P): orange
⬤ Nitrogen (N): blue
⬤ Selenium (Se): gold
⬤ Sulfur (S): yellow
Summary
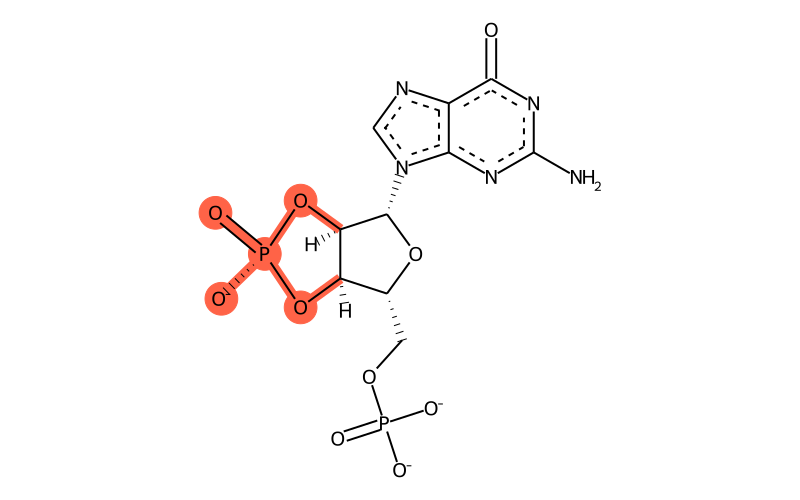
| Full name | guanoside-5'-phosphate-2',3'-cyclic phosphate |
| Short name | pG2'3'cp |
| MODOMICS code new | 2000003377G |
| MODOMICS code | 3377G |
| Nature of the modified residue | Natural |
| Residue unique ID | 238 |
| Found in RNA | Yes |
| Related nucleosides | 38 |
| RCSB ligands |
Chemical information
| Sum formula | C10H9N5O10P2 |
| Type of moiety | nucleotide |
| Degeneracy | not aplicable |
| SMILES | [C@H]1([n]2cnc3c2nc(nc3=O)N)[C@@H]2O[P@]([O-])(=O)O[C@@H]2[C@@H](COP([O-])([O-])=O)O1 |
| logP | -1.9061 |
| TPSA | 243.27 |
| Number of atoms | 27 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 12 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 15 |
| Number of Hydrogen Bond Donors (HBD) | 1 |
| InChI | InChI=1S/C10H12N5O10P2/c11-10-13-7-4(8(16)14-10)12-2-15(7)9-6-5(24-27(20,21)25-6)3(23-9)1-22-26(17,18)19/h2-3,5-6,9H,1H2,(H,20,21)(H2,11,14,16)(H2,17,18,19)/p-3/t3-,5-,6-,9-/m1/s1 |
| InChIKey | MYDHAVOCUFAOHX-UUOKFMHZSA-K |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 29.966535453 |
| Energy [Eh]: | -2095.69348519787 |
| HOMO [eV]: | -8.3322 |
| LUMO [eV]: | 1.5068 |
| Gap [eV]: | 9.839 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
LC-MS Information
| Monoisotopic mass | None |
| Average mass | 421.153 |
| [M+H]+ | not available |
| Product ions | not available |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Last modification of this entry: Sept. 15, 2025