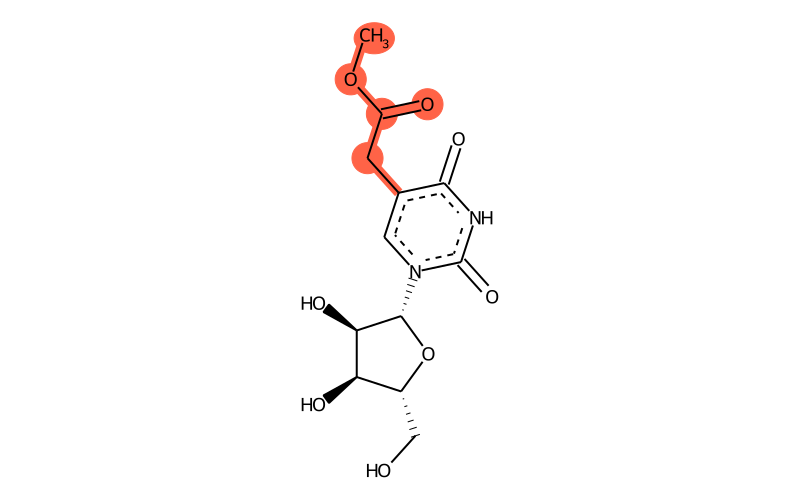
Summary
| Full name | 5-methoxycarbonylmethyluridine |
| IUPAC name | methyl 2-[1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2,4-dioxopyrimidin-5-yl]acetate |
| Short name | mcm5U |
| MODOMICS code new | 2000000521U |
| MODOMICS code | 521U |
| Synonyms |
1,2,3,4-Tetrahydro-2,4-dioxo-1-beta-D-ribofuranosyl-5-pyrimidineacetic Acid Methyl Ester
29428-50-0 5-(2-methoxy-2-oxoethyl)uridine 5-(2-Oxo-2-methoxyethyl)uridine 5-Mcmu 5-Methoxycarbonyl Methyl Uridine 5-Methoxycarbonylmethyluridine 5-(methoxycarbonyl)methyluridine 5-Pyrimidineacetic acid, 1,2,3,4-tetrahydro-2,4-dioxo-1-beta-D-ribofur anosyl-, methyl ester 5-Pyrimidineacetic acid, 1,2,3,4-tetrahydro-2,4-dioxo-1-beta-D-ribofuranosyl-, methyl ester 5-Pyrimidineaceticacid, 1,2,3,4-tetrahydro-2,4-dioxo-1-b-D-ribofuranosyl-, methyl ester 5-Pyrimidineaceticacid,1,2,3,4-tetrahydro-2,4-dioxo-1-beta-D-ribofuranosyl-,methylester AC1MIVNC C12-H16-N2-O8 C12H16N2O8 CHEBI:20598 CID3080753 DTXSID20183642 imidin-5-yl)acetate mcm(5)u mcm5U methyl 2-[1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2,4-dioxopyrimidin-5-yl]acetate methyl 2-(1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2,4-dioxo-1,2,3,4-tetrahydropyr Methyl 2-(1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)acetate methyl uridine 5-acetate Q27109316 SCHEMBL64345 uridine 5-acetic acid methyl ester YIZYCHKPHCPKHZ-PNHWDRBUSA-N ZINC33902395 |
| Nature of the modified residue | Natural |
| RNAMods code | 1 |
| Residue unique ID | 27 |
| Found in RNA | Yes |
| Related nucleotides | 362 |
| Enzymes |
ALKBH8 (Homo sapiens) TRM112 (Homo sapiens) Trm9 (Saccharomyces cerevisiae) Trm9 (Haloferax volcanii) |
| Found in phylogeny | Eukaryota |
| Found naturally in RNA types | tRNA |
Chemical information
| Sum formula | C12H16N2O8 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| ChEBI ID | 20598 |
| CAS Registry Number | 29428-50-0 |
| Reaxys Registry Number | 964573 |
| SMILES | COC(Cc1c(=O)[nH]c(=O)[n]([C@H]2[C@H](O)[C@H](O)[C@@H](CO)O2)c1)=O |
| logP | -3.1364 |
| TPSA | 151.08 |
| Number of atoms | 22 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C12H16N2O8/c1-21-7(16)2-5-3-14(12(20)13-10(5)19)11-9(18)8(17)6(4-15)22-11/h3,6,8-9,11,15,17-18H,2,4H2,1H3,(H,13,19,20)/t6-,8-,9-,11-/m1/s1 |
| InChIKey | YIZYCHKPHCPKHZ-PNHWDRBUSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
9470984
36411681 57354413 79023203 111681181 129802761 135227312 135668179 224671896 226445609 241049882 252228132 252419320 260378153 273111440 312237547 315744021 319287649 340590054 341246955 348953990 355132847 375460337 377951602 385664025 386500541 388915100 404623038 419579747 439384254 441133767 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 6.64363726 |
| Energy [Eh]: | -1177.80755187584 |
| HOMO [eV]: | -9.3654 |
| LUMO [eV]: | 0.725 |
| Gap [eV]: | 10.0904 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
COC(C=C1C(=O)NC(=O)N(C2C(O)C(O)C(CO)O2)C1)=O tautomer #0
COC(Cc1c(=O)[nH]c(=O)n(C2C(O)C(O)C(CO)O2)c1)=O tautomer #1 COC(=Cc1c(=O)[nH]c(=O)n(C2C(O)C(O)C(CO)O2)c1)O tautomer #2 COC(C=C1C(O)=NC(=O)N(C2C(O)C(O)C(CO)O2)C1)=O tautomer #3 COC(Cc1c(O)nc(=O)n(C2C(O)C(O)C(CO)O2)c1)=O tautomer #4 COC(C=C1C(=O)N=C(O)N(C2C(O)C(O)C(CO)O2)C1)=O tautomer #5 COC(Cc1c(=O)nc(O)n(C2C(O)C(O)C(CO)O2)c1)=O tautomer #6 COC(=Cc1c(O)nc(=O)n(C2C(O)C(O)C(CO)O2)c1)O tautomer #7 COC(=Cc1c(=O)nc(O)n(C2C(O)C(O)C(CO)O2)c1)O tautomer #8 |
| Tautomer image | Show Image |
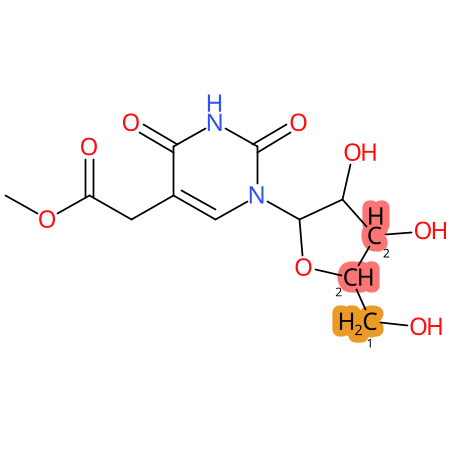
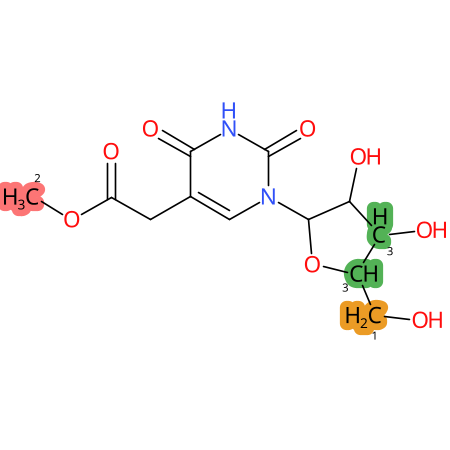
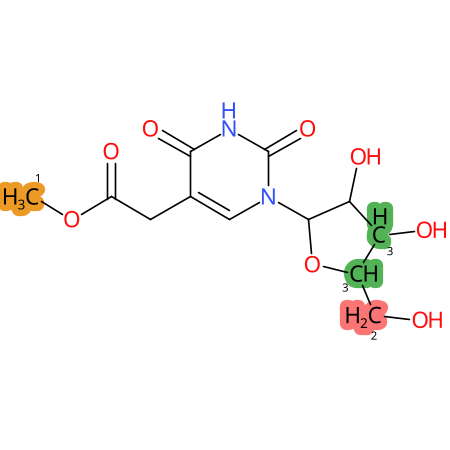
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 316.0907 |
| Average mass | 316.264 |
| [M+H]+ | 317.0985 |
| Product ions | 185/153/125 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| other | carboxymethyl |
| other | methylester |
Reactions producing 5-methoxycarbonylmethyluridine
| Name |
|---|
| cm5U:mcm5U |
Reactions starting from 5-methoxycarbonylmethyluridine
| Name |
|---|
| mcm5U:mchm5U |
| mcm5U:mcm5s2U |
| mcm5U:mcm5Um |
Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons | Sakai Y, Miyauchi K, Kimura S, Suzuki T. | Nucleic Acids Res. | [details] | 26681692 | 10.1093/nar/gkv1470 |
Last modification of this entry: Sept. 15, 2025