Summary
| Full name | N6,N6-dimethyladenosine |
| IUPAC name | (2R,3R,4S,5R)-2-[6-(dimethylamino)purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol |
| Short name | m6,6A |
| MODOMICS code new | 2000000066A |
| MODOMICS code | 66A |
| Synonyms |
0L9766EW9W
1164102-97-9 2620-62-4 26A (2R,3R,4S,5R)-2-(6-(Dimethylamino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (2R,3R,4S,5R)-2-[6-(dimethylamino)purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol (2R,3R,4S,5R)-2-[6-(dimethylamino)purin-9-yl]-5-(hydroxymethyl)tetrahydrofuran-3,4-diol 6-Dimethyladenosine 6-Dimethylamino-9-(b-D-ribofuranosyl)purine 6-Dimethylaminopurine 9-riboside 6-dimethylaminopurine 9-riboside*crystalline 6-Dimethylaminopurine D-riboside 6-(Dimethylamino)purine ribonucleoside 6-dimethylaminopurine riboside 6-(Dimethylamino)purine riboside 6-Dimethylaminopurine-9-riboside 6-(Dimethylamino)purineribonucleoside 6-(gamma,gamma-dimethylamino)purine riboside 6-N-Dimethyladenosine 6N-DIMETHYLADENOSINE AC1L98HI Adenosine, N,N-dimethyl- Adenosine,N,N-dimethyl- AKOS027327106 BS-27732 C03416 C12-H17-N5-O4 C12H17N5O4 CHEBI:28284 CHEMBL1229848 CID440004 CS-0022419 DTXSID90949075 EINECS 220-057-7 Epitope ID:140944 HY-101984 MFCD00022822 N6-Dimethyladenosine n6,n6-dimethyl-adenosine N6,N6-Dimethyladenosine N(6),N(6)-Dimethyladenosine N/A N,N-Dimethyl-9-pentofuranosyl-9H-purin-6-amine N,N-Dimethyladenosine NS00045585 NSC 627046 NSC627046 Q15632696 SCHEMBL157530 UNII-0L9766EW9W WVGPGNPCZPYCLK-WOUKDFQISA-N ZINC4096103 |
| Nature of the modified residue | Natural |
| RNAMods code | ζ |
| Residue unique ID | 41 |
| Found in RNA | Yes |
| Related nucleotides | 192 |
| Enzymes |
DIMT1 (Caenorhabditis elegans) RsmA (Escherichia coli) RsmA (Saccharomyces cerevisiae) RsmA (Thermus thermophilus) RsmA (Pseudomonas aeruginosa) TFB1M (Mus musculus) ksgA (Haloferax volcanii) mtTFBp (Saccharomyces cerevisiae) |
| Found in phylogeny | Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA |
Chemical information
| Sum formula | C12H17N5O4 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 72266 |
| ChEBI ID | 28284 |
| CAS Registry Number | 2620-62-4 |
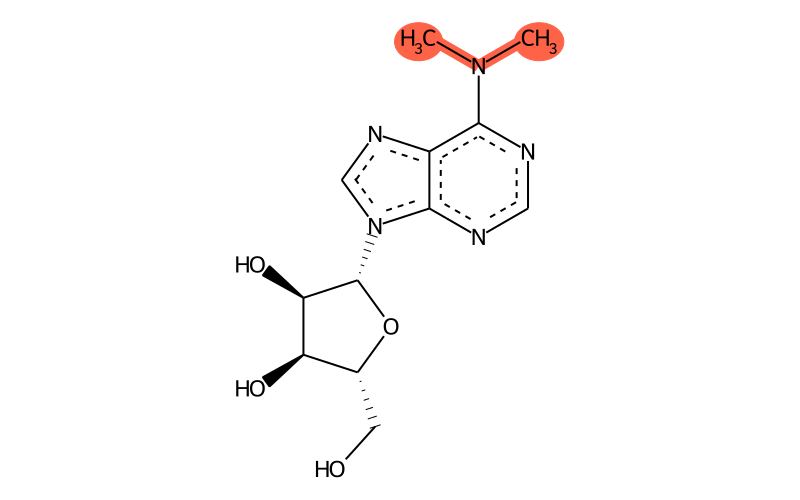
| SMILES | CN(C)c1ncnc2c1nc[n]2[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
| logP | -1.4962 |
| TPSA | 116.76 |
| Number of atoms | 21 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| InChI | InChI=1S/C12H17N5O4/c1-16(2)10-7-11(14-4-13-10)17(5-15-7)12-9(20)8(19)6(3-18)21-12/h4-6,8-9,12,18-20H,3H2,1-2H3/t6-,8-,9-,12-/m1/s1 |
| InChIKey | WVGPGNPCZPYCLK-WOUKDFQISA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
6245
597012 10298511 14710000 14710003 14775888 36884134 79284888 85164663 87895529 104246043 104621978 129639276 135030135 140033852 143178691 175442861 188889072 226521399 241057767 250011763 252165601 252400604 252400657 257139667 275185433 277393114 310272021 312236038 314983140 317882586 319221882 319483675 341261213 346763231 347730739 348202442 348935704 355136785 374044427 374372481 375111170 375300395 377532169 378032239 381318859 381833099 381833338 384506116 385644306 386480622 387171983 389769234 419585666 435888463 439368918 439446460 441168994 442087268 442834470 442877580 442907526 443529900 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 3.896162378 |
| Energy [Eh]: | -1041.80768751479 |
| HOMO [eV]: | -8.4023 |
| LUMO [eV]: | 1.1328 |
| Gap [eV]: | 9.5351 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
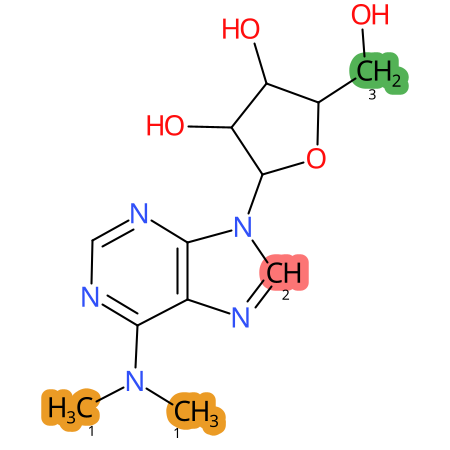
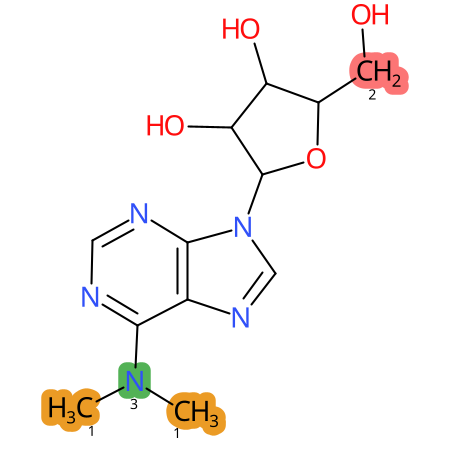
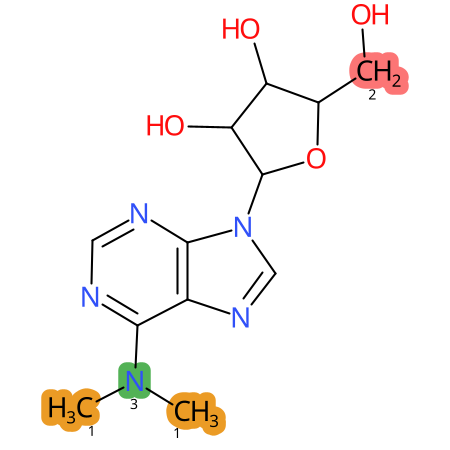
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 295.1281 |
| Average mass | 295.294 |
| [M+H]+ | 296.1359 |
| Product ions | 164 |
| Normalized LC elution time * | 1,86 (Kellner 2014); 2,57 (Kellner 2014) |
| LC elution order/characteristics | after m6A (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at other N |
| methyl group | second methyl at other N |
Reactions producing N6,N6-dimethyladenosine
| Name |
|---|
| m6A:m6,6A |
Reactions starting from N6,N6-dimethyladenosine
| Name |
|---|
| m6,6A:m6,6Am |
Last modification of this entry: Sept. 15, 2025