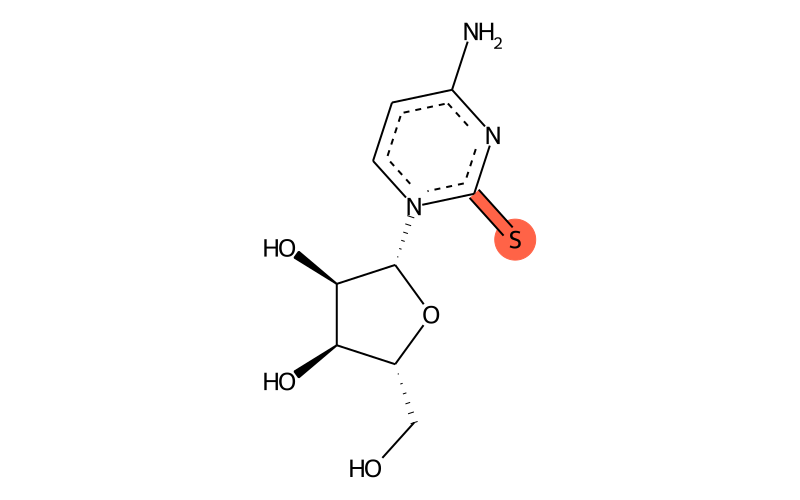
Summary
| Full name | 2-thiocytidine |
| IUPAC name | 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2-thione |
| Short name | s2C |
| MODOMICS code new | 2000000002C |
| MODOMICS code | 2C |
| Synonyms |
13239-97-9
2(1H)-Pyrimidinethione, 4-amino-1-.beta.-D-ribofuranosyl- 2-Thiocytidine 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2-thione 4-Amino-1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)pyrimidine-2(1H)-thione 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidine-2-thione 4-amino-1-beta-D-ribofuranosylpyrimidine-2(1H)-thione AC1MHUQ1 beta-D-ribofuranosyl-2-thiocytidine BRD-K67390410-001-01-2 C9-H13-N3-O4-S C9H13N3O4S CHEBI:19780 CID3011746 CTK0H9573 Cytidine, 2-thio- DTXSID30157516 EINECS 236-214-8 N/A NS00024211 Q20890505 SCHEMBL42103 ZINC13516196 |
| Nature of the modified residue | Natural |
| RNAMods code | ʤ |
| Residue unique ID | 92 |
| Found in RNA | Yes |
| Related nucleotides | 347 |
| Enzymes |
TtcA (Escherichia coli) |
| Found in phylogeny | Eubacteria |
| Found naturally in RNA types | tRNA |
Chemical information
| Sum formula | C9H13N3O4S |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 3011746 |
| ChEBI ID | 19780 |
| CAS Registry Number | 13239-97-9 |
| SMILES | Nc1nc(=S)[n]([C@H]2[C@H](O)[C@H](O)[C@@H](CO)O2)cc1 |
| logP | -0.6125 |
| TPSA | 145.85 |
| Number of atoms | 17 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 7 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 8 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C9H13N3O4S/c10-5-1-2-12(9(17)11-5)8-7(15)6(14)4(3-13)16-8/h1-2,4,6-8,13-15H,3H2,(H2,10,11,17)/t4-,6-,7-,8-/m1/s1 |
| InChIKey | RHFUOMFWUGWKKO-XVFCMESISA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
3724289
10039639 14917406 29214843 36060001 57410215 75677304 111632952 129129091 135041362 160657135 162475556 226426961 249759512 252333563 252400680 259323547 274890304 310280892 312237517 315718034 318488395 319249902 341111164 341246632 349636061 355099528 363671150 375198028 375553690 381028159 381998526 385541238 385647743 386484149 387148260 403601584 419580118 434400811 439476913 442933857 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 13.352177515 |
| Energy [Eh]: | -1213.73445195662 |
| HOMO [eV]: | -8.5705 |
| LUMO [eV]: | 0.6676 |
| Gap [eV]: | 9.2381 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
N=c1[nH]c(=S)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #0
N=c1nc(S)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #1 Nc1nc(=S)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #2 |
| Tautomer image | Show Image |
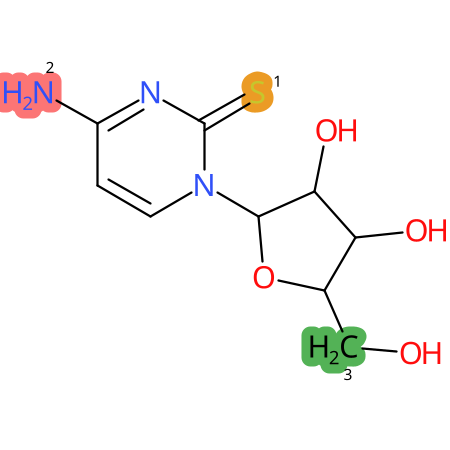
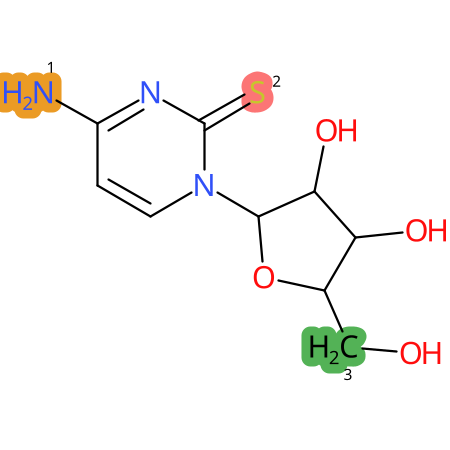
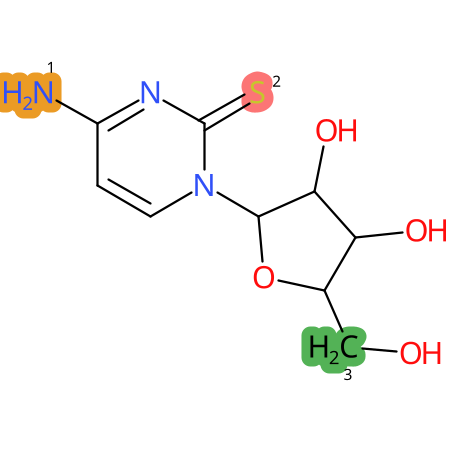
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 259.0627 |
| Average mass | 259.282 |
| [M+H]+ | 260.0705 |
| Product ions | 128 |
| Normalized LC elution time * | 0,68 (Kellner 2014); 0,86 (Kellner 2014) |
| LC elution order/characteristics | between U and G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Comments
s2C at position 32 in bacterial tRNA allows more anticodon loop flexibility that limit formation of the characteristic U33-turn conformation of anticodon loop in other tRNA lacking s2C32.
Chemical groups contained
| Type | Subtype |
|---|---|
| heavy atom | sulfur |
Reactions producing 2-thiocytidine
| Name |
|---|
| C:s2C |
Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Modifications Modulate Anticodon Loop Dynamics and Codon Recognition of E. coli tRNA(Arg1,2). | Cantara WA, Bilbille Y, Kim J, Kaiser R, Leszczyńska G, Malkiewicz A, Agris PF | J Mol Biol | [details] | 22240457 | - |
| Thiobases in Escherchia coli Transfer RNA: 2-Thiocytosine and 5-Methylaminomethyl-2-thiouracil. | Carbon J, David H, Studier MH | Science | [details] | 17812290 | - |
Last modification of this entry: Sept. 15, 2025