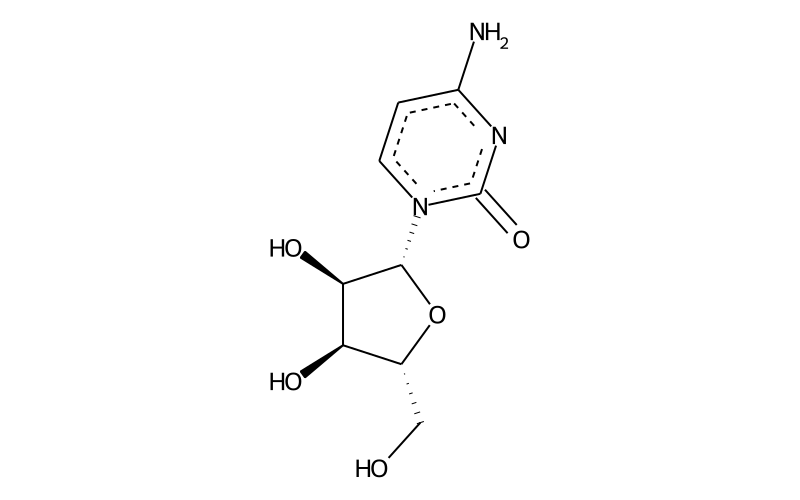
Summary
| Full name | cytidine |
| IUPAC name | 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one |
| Short name | C |
| MODOMICS code new | C |
| MODOMICS code | C |
| Synonyms |
111398-92-6
14419-78-4 147-94-4 16501-EP2281563A1 16501-EP2287165A2 16501-EP2287166A2 16501-EP2289892A1 16501-EP2292088A1 16501-EP2292620A2 16501-EP2292630A1 16501-EP2295407A1 16501-EP2298736A1 16501-EP2298783A1 16501-EP2305808A1 16501-EP2316459A1 185553-94-0 1beta-2'-Ribofuranosylcytosine, d- 1-Beta-d-ribofuranoside cytosine 1-(beta-D-Ribofuranosyl)-2-oxo-4-amino-1,2-dihydro-1,3-diazine 1-(beta-D-ribofuranosyl)-4-aminopyrimidin-2-one 1-beta-D-Ribofuranosylcytosine 1.beta.-D-Ribofuranosylcytosine 1beta-D-Ribofuranosylcytosine 1-beta-D-ribosyl- (6CI) 1-beta-Ribofuranosylcytosine 1.beta.-Ribofuranosylcytosine 1beta-Ribofuranosylcytosine 2(1H)-Pyrimidinone, 4-amino-1-beta-D-ribofuranosyl- 2(1H)-Pyrimidinone, 4-amino-1-.beta.-D-ribofuranosyl- 2(1H)-Pyrimidinone, 4-amino-1beta-D-ribofuranosyl- 32747-18-5 3h-cytidine 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2-dihydropyrimidin-2-one 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidin-2-one 4-amino-1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)pyrimidin-2(1H)-one 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidin-2-one 4-Amino-1-beta-D-ribofuranosyl-2(1H)-pyrimidinone 4-Amino-1-beta-D-ribofuranosyl-2-(1H)-pyrimidinone 4-Amino-1.beta.-D-ribofuranosyl-2(1H)-pyrimidinone 4-Amino-1-.beta.-D-ribofuranosyl-2-(1H)-pyrimidinone 4-Amino-1beta-D-ribofuranosyl-2(1H)-pyrimidinone 4-amino-1-beta-D-ribofuranosylpyrimidin-2(1H)-one 5CSZ8459RP 6018-48-0 65-46-3 65C463 67875-14-3 6D2DC474-DD76-4081-8B34-10605C218F49 AB0012341 AC1L1LY9 AC1Q6C2P ACN-047006 AJ-43263 AKOS015888568 AM83932 AN-23747 ANW-35046 AR-1I3421 AS-12696 BC679398 beta-cytidine beta.-D-Ribo-C .beta.-D-Ribofuranoside, cytosine-1 beta-D-Ribofuranoside, cytosine-1 beta-D-ribofuranosyl-cytidine bmse000190 bmse000969 bmse001020 BRD-K71847383-001-12-5 C00475 c0522 C9-H13-N3-O5 C9H13N3O5 CCG-266896 CHEBI:17562 CHEMBL95606 CID6175 CJ-10310 CP-C CS-1989 CTK2F3027 CTN Cyd CYT007 cytarabine cytidin- Cytidin cytidine Cytidine- Cytidine, 99% Cytidine, 99+% Cytidine, 99% - 100g 100g Cytidine 99% for biochemistry 1gm Cytidine, >=99.0% (HPLC) Cytidine, BioReagent, suitable for cell culture, powder, >=99% cytidine, N-15N- cytidine-2'-13C Cytidine-5'-monophosphate cytosine Cytosine, 1-beta-D-ribofuranosyl- Cytosine, 1-.beta.-D-ribofuranosyl- Cytosine beta-D-riboside Cytosine Ribonucleoside Cytosine riboside Cytosine-1-beta-D-ribofuranoside cytosine-1beta-D-Ribofuranoside Cytosine-1-??-D-ribofuranoside D003562 D07769 D0E7ES DB02097 DB-029614 DTXSID60891552 EINECS 200-610-9 Epitope ID:141494 ERK5-5D16 F0348-2240 GTPL4728 HY-B0158 I14-8299 J-700167 KM0929 KSC353A2P LS-135845 MCULE-3044641322 MFCD00006545 MLS000049947 MLS002207040 MMV638723 NCGC00093356-08 NCGC00142483-01 NCGC00142483-09 NS00069068 NSC 20258 NSC-20258 Posilent (TN) PubChem14146 Q422538 Ribonucleoside, Cytosine Riboside, Cytosine RTR-022281 s2053 SBB003195 SC-09086 SCHEMBL7179 SMR000058243 SRI-2352_17 ST056938 ST24046405 TL8004649 TR-022281 UHDGCWIWMRVCDJ-XVFCMESISA-N UNII-5CSZ8459RP UNII-F2 component UHDGCWIWMRVCDJ-XVFCMESISA-N ZINC2583632 Zytidin |
| Nature of the modified residue | Natural |
| RNAMods code | C |
| Residue unique ID | 46 |
| Found in RNA | Yes |
| Related nucleotides | 300 |
| Enzymes |
ALKBH3 (Homo sapiens) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
Chemical information
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
N=c1[nH]c(=O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #0
Nc1nc(=O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #1 N=c1nc(O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #2 |
| Tautomer image | Show Image |
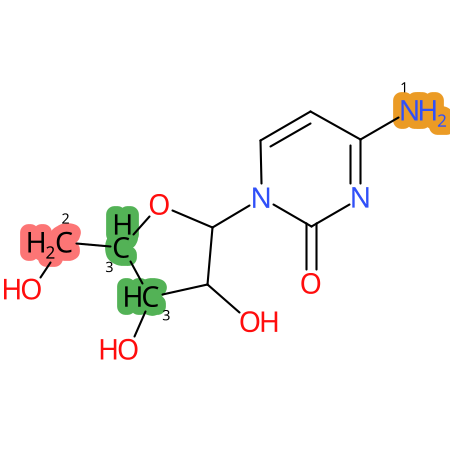
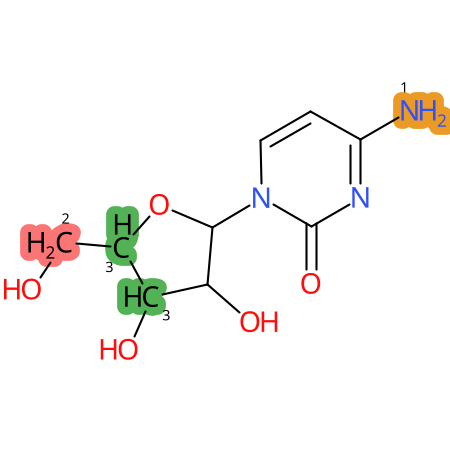
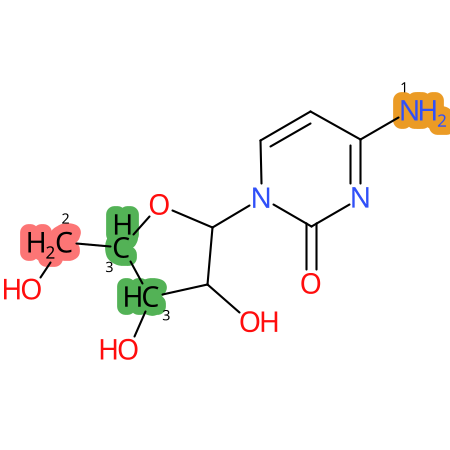
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 243.0855 |
| Average mass | 243.217 |
| [M+H]+ | 244.0933 |
| Product ions | 112 |
| Normalized LC elution time * | 0.42 |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Reactions producing cytidine
| Name |
|---|
| m3C:C |
Reactions starting from cytidine
| Name |
|---|
| C:k2C |
| C:ac4C |
| C:m3C |
| C:f5C |
| C:Cm |
| C:m4C |
| C:m5C |
| C:hm5C |
| C:s2C |
| C:U |
| C:C+ |
| C:ho5C |
Last modification of this entry: Oct. 19, 2023