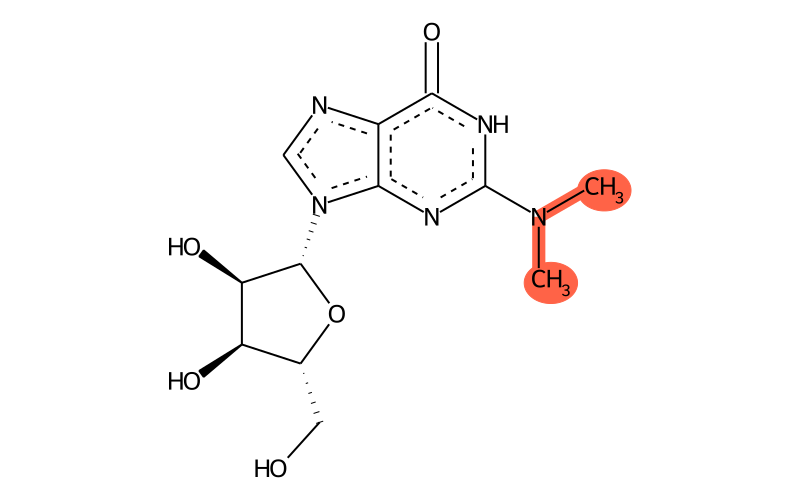
Summary
| Full name | N2,N2-dimethylguanosine |
| IUPAC name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-1H-purin-6-one |
| Short name | m2,2G |
| MODOMICS code new | 2000000022G |
| MODOMICS code | 22G |
| Synonyms |
0881AB
13269-19-7 2- GUANOSINE 2140-67-2 2,2-Dimethylguanosine 2-Dimethylamino-6-oxypurine riboside 2-Dimethylamino-6-oxypurineriboside 2-(dimethylamino)-9-(beta-D-ribofuranosyl)-1,9-dihydro-6H-purin-6-one 2-(Dimethylamino)-9-pentofuranosyl-9H-purin-6-ol 2-Dimethylaminoguanosine 2-(DIMETHYLAMINO)GUANOSINE 2-Dimethylguanosine 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-3H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-6,9-dihydro-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(dimethylamino)-6,9-dihydro-3H-purin-6-one 9-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2-(dimethylamino)-1H-purin-6(9H)-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-2-(dimethylamino)-1H-purin-6-one 9-((2S,3S,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)-tetrahydrofuran-2-yl)-2-(dimethylamino)-1H-purin-6(9H)-one AC1L3P3V AJ-53133 AKOS016009587 AR-1K2505 AS-57489 C12H17N5O5 CHEBI:19289 CID92919 CS-0061838 CTK4E6678 Dimethylguanosine DTXSID90943957 Guanosine, N,N-dimethyl- Guanosine,N,N-dimethyl- HY-113137 m22g MFCD00057049 N2-Dimethylguanosine N2,N2-Dimethylguanosin N2,N2-Dimethylguanosine N(2),N(2)-Dimethylguanosine N,N- dimethylguanosine N,N-dimethyl-Guanosine N,N-Dimethylguanosine NS00099785 P7NB8Q3W0R Q27109160 RSPURTUNRHNVGF-IOSLPCCCSA-N SCHEMBL40775 UNII-P7NB8Q3W0R ZINC5115341 |
| Nature of the modified residue | Natural |
| RNAMods code | R |
| Residue unique ID | 13 |
| Found in RNA | Yes |
| Related nucleotides | 198 |
| Enzymes |
Trm-G10 (Pyrococcus abyssi) Trm1 (Saccharomyces cerevisiae) Trm1 (Aquifex aeolicus) Trm1 (Pyrococcus horikoshii) Trm1 (Homo sapiens) trmG10 (Haloferax volcanii) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, tRNA |
Chemical information
| Sum formula | C12H17N5O5 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 601633 |
| ChEBI ID | 19289 |
| CAS Registry Number | 2140-67-2 |
| Reaxys Registry Number | 47545 |
| SMILES | CN(C)c1[nH]c(=O)c2nc[n]([C@@H]3O[C@H](CO)[C@@H](O)[C@H]3O)c2n1 |
| logP | -2.2029 |
| TPSA | 136.73 |
| Number of atoms | 22 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C12H17N5O5/c1-16(2)12-14-9-6(10(21)15-12)13-4-17(9)11-8(20)7(19)5(3-18)22-11/h4-5,7-8,11,18-20H,3H2,1-2H3,(H,14,15,21)/t5-,7-,8-,11-/m1/s1 |
| InChIKey | RSPURTUNRHNVGF-IOSLPCCCSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
15975949
17399061 44423698 50500055 53801190 57335432 89097505 103836704 104408712 129175906 135052147 162814995 163658117 163848499 171579067 223562304 226425943 241043528 250058384 252400658 252462248 255436963 257599240 275189595 275918333 297451248 310272006 312226815 313053461 314981618 319475695 341143147 342526832 347744790 347752891 348276386 348902047 355088131 363671324 373765433 375091221 375539897 375973960 375987067 375987318 376149742 376512219 378023108 384232301 384432628 384501007 385663819 386500335 387024011 387150448 389289958 404769319 404825307 419580279 433774886 433775448 434369768 434407460 436398348 440820846 442031543 442860983 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 11.061397064 |
| Energy [Eh]: | -1117.02274734863 |
| HOMO [eV]: | -8.2207 |
| LUMO [eV]: | 1.4609 |
| Gap [eV]: | 9.6816 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
CN(C)c1nc(O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #0
CN(C)c1nc(=O)c2ncn(C3OC(CO)C(O)C3O)c2[nH]1 tautomer #1 CN(C)c1[nH]c(=O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #2 CN(C)c1nc(O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #3 CN(C)C1=NC(=O)C2N=CN(C3OC(CO)C(O)C3O)C2=N1 tautomer #4 |
| Tautomer image | Show Image |
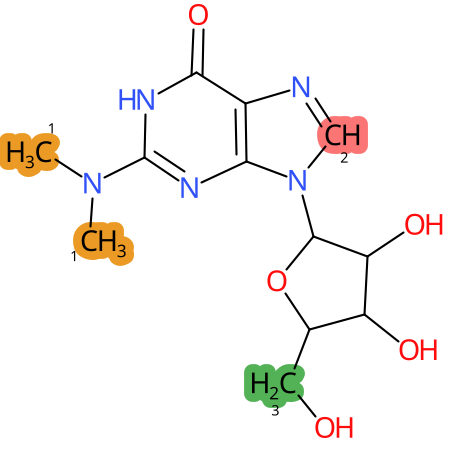
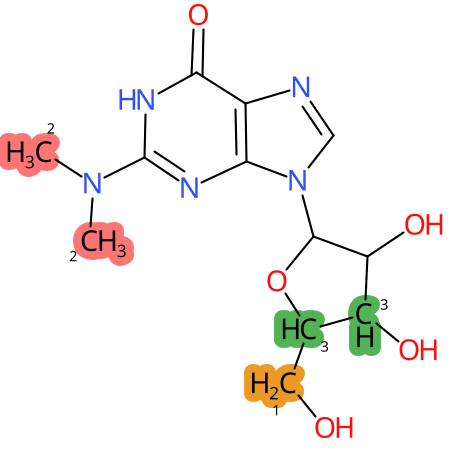
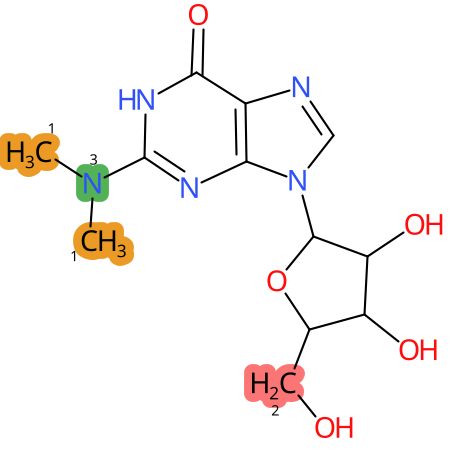
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 311.123 |
| Average mass | 311.294 |
| [M+H]+ | 312.1308 |
| Product ions | 180 |
| Normalized LC elution time * | 1,62 (Kellner 2014) |
| LC elution order/characteristics | between A and m6A (Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at other N |
| methyl group | second methyl at other N |
Reactions producing N2,N2-dimethylguanosine
| Name |
|---|
| m2G:m2,2G |
Reactions starting from N2,N2-dimethylguanosine
| Name |
|---|
| m2,2G:m2,2Gm |
Last modification of this entry: Sept. 15, 2025