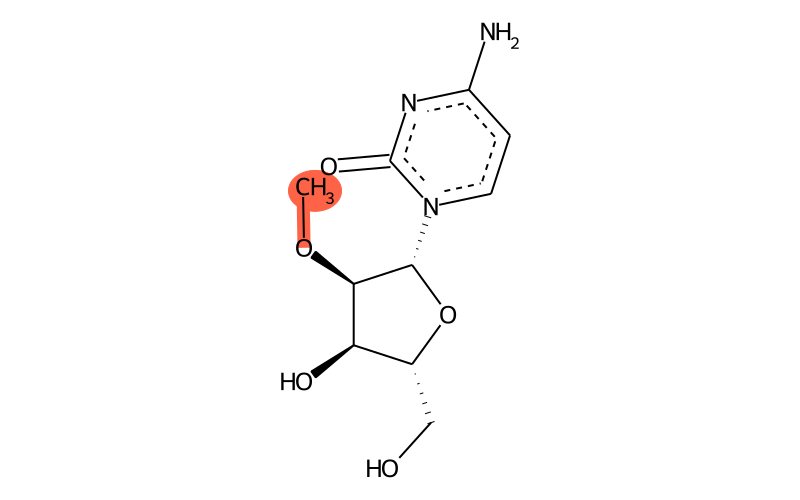
Summary
| Full name | 2'-O-methylcytidine |
| IUPAC name | 4-amino-1-[(2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxyoxolan-2-yl]pyrimidin-2-one |
| Short name | Cm |
| MODOMICS code new | 2000000090C |
| MODOMICS code | 0C |
| Synonyms |
2140-72-9
2NULL-O-Methylcytidine 2'-O-Methyl cytidine 2''-O-Methylcytidine 2'-O-Methylcytidine 2/'-O-Methylcytidine 2'-O-METHYL-CYTIDINE 2-O-METHYLCYTIDINE 1G 2-O-METHYLCYTIDINE 200MG 2'-O-Methylcytidine =98.0% (by HPLC, titration analysis) 2'-O-Methylcytidine, 99% - 1G 1g 4-amino-1-[(2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxyoxolan-2-yl]pyrimidin-2-one 4-amino-1-((2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxytetrahydrofuran-2-yl)pyrimidin-2(1H)-one 4-amino-1-[(2R,3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)-3-methoxy-tetrahydrofuran-2-yl]pyrimidin-2-one 4-Amino-1-((2R,3R,4S,5R)-4-hydroxy-5-hydroxymethyl-3-methoxy-tetrahydro-furan-2-yl)-1H-pyrimidin-2-one 4-Imino-1-(2-O-methylpentofuranosyl)-1,4-dihydropyrimidin-2-ol 58970-16-4 5XD9IH2G27 AB0011995 AC1L45EF AIDS338901 AJ-55451 AK-54700 AKOS015850975 AKOS015896935 ANW-54599 BRD-K23276845-001-01-1 C10H15N3O5 CHEBI:19228 CHEMBL176605 CID150971 CJ-12849 CM-782 CTK1A1816 Cytidine, 2'-O-methyl- DS-14587 DTXSID50943958 M-3929 MFCD00056067 O-2-Methylcytidine O(2')-Methylcytidine O2'-Methylcytidine PubChem11055 Q15632793 RFCQJGFZUQFYRF-ZOQUXTDFSA-N RP29164 SC-23539 SCHEMBL64632 TC-071340 TL8001776 UNII-5XD9IH2G27 ZB1481 ZINC5998209 |
| Nature of the modified residue | Natural |
| RNAMods code | B |
| Residue unique ID | 84 |
| Found in RNA | Yes |
| Related nucleotides | 201 |
| Enzymes |
Nop1 (Saccharomyces cerevisiae) RlmM (Escherichia coli) RsmI (Escherichia coli) Trm13 (Saccharomyces cerevisiae) Trm7 (Saccharomyces cerevisiae) TrmJ (Escherichia coli) TrmJ (Sulfolobus acidocaldarius) TrmL (Escherichia coli) aTrm56 (Pyrococcus abyssi) |
| Found in phylogeny | Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, snRNA, tRNA |
Chemical information
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
COC1C(n2ccc(=N)[nH]c2=O)OC(CO)C1O tautomer #0
COC1C(n2ccc(N)nc2=O)OC(CO)C1O tautomer #1 COC1C(n2ccc(=N)nc2O)OC(CO)C1O tautomer #2 |
| Tautomer image | Show Image |
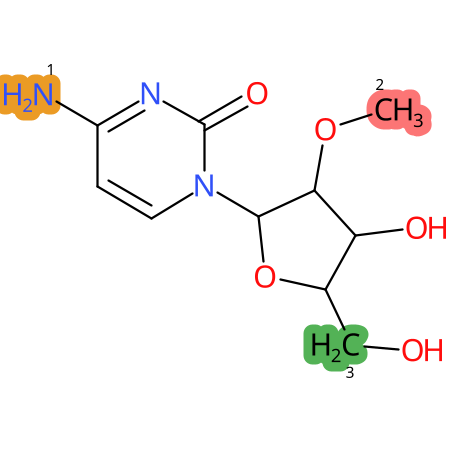
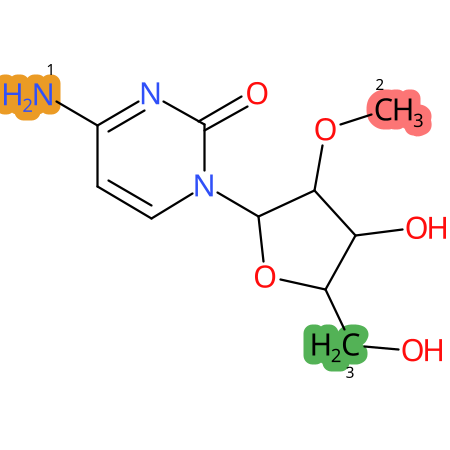
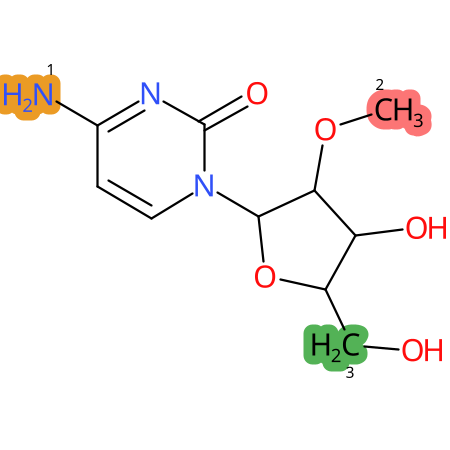
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 257.1012 |
| Average mass | 257.243 |
| [M+H]+ | 258.1082 |
| Product ions | 112 |
| Normalized LC elution time * | 0,90 (Kellner 2014); 0,92 (Kellner 2014) |
| LC elution order/characteristics | between U and G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at O2 |
Reactions producing 2'-O-methylcytidine
| Name |
|---|
| C:Cm |
Reactions starting from 2'-O-methylcytidine
| Name |
|---|
| Cm:m4Cm |
| Cm:m5Cm |
| Cm:ac4Cm |
| Cm:f5Cm |
Last modification of this entry: Sept. 22, 2023