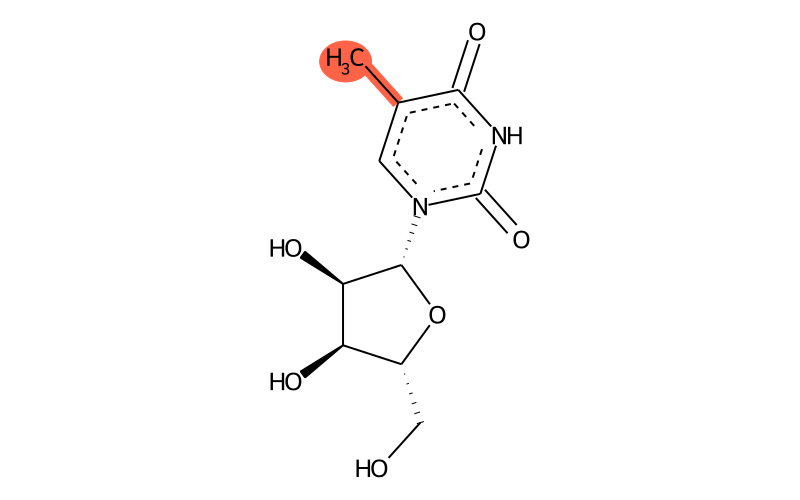
Summary
| Full name | 5-methyluridine |
| IUPAC name | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidine-2,4-dione |
| Short name | m5U |
| MODOMICS code new | 2000000005U |
| MODOMICS code | 5U |
| Synonyms |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)-2-oxolanyl]-5-methylpyrimidine-2,4-dione
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2,3,4-tetrahydropyrimidine-2,4-dione 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidine-2,4-dione 1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-5-methyl-pyrimidine-2,4-dione 1-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione 1-((2R,3R,4S,5R)-TETRAHYDRO-3,4-DIHYDROXY-5-(HYDROXYMETHYL)FURAN-2-YL)-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE 1463-10-1 1-[(4S,2R,3R,5R)-3,4-Dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,3-dihydropyrimidine-2,4-dione 172163-66-5 180000-19-5 1-b-D-Ribofuranosylthymine 1-beta-delta-Ribofuranosylthymine 1-(beta-D-ribofuranosyl)thymine 1-.beta.-D-Ribofuranosylthymine 2,4(1H,3H)-Pyrimidinedione, 5-methyl-1-.beta.-D-ribofuranosyl- 38T 463M101 5-methyl-1-beta-delta-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione 5-methyl-1-beta-D-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione 5-methyl-Uridine 5-Methyluridine 5-METHYLURIDINE 5'-MONOPHOSPHATE 5-Methyluridine, 97% 5-Methyluridine, 99% - 25G 25g 5-Methyluridine, powder 5MU 5MU-5MU-5MU A808481 AC1L9HXX AC-8172 ACT03080 AJ-43265 AKOS015896830 AM83947 ANW-21004 Arabinosylthymine b-D-Ribofuranoside thymine-1 beta-delta-Ribofuranoside beta-delta-Ribofuranoside thymine-1 beta-D-Ribofuranoside .beta.-D-Ribofuranoside, thymine-1 .beta.-D-Ribofuranosylthymine bmse000759 C10-H14-N2-O6 C10H14N2O6 CHEBI:30821 CHEBI:45996 CHEMBL106175 CID445408 CJ-10312 CM0074 CS-W010160 CTK0H4640 DB-030334 DS-18476 DTXSID20163348 DWRXFEITVBNRMK-JXOAFFINSA-N EINECS 215-973-9 Epitope ID:141497 HY-W009444 I14-2705 J-700101 KS-00000NGX KSC174M4B M1405 MFCD00006535 NS00024786 PubChem14166 Q425078 ribosylthymidine Ribosylthymine Ribothymidine RL01838 s6156 SC-09124 SCHEMBL29298 ST2406328 STL507242 Thymine ribofuranoside Thymine ribonucleoside Thymine riboside thymine-1 beta-delta-Ribofuranosylthymine thymine-1 beta-D-Ribofuranosylthymine TL8001022 UNII-ZS1409014A Uridine, 5-methyl- uridine-1'-14C-5-methyl- uridine-1,3-15N2-5-methyl- ZINC2583634 ZS1409014A |
| Nature of the modified residue | Natural |
| RNAMods code | T |
| Residue unique ID | 12 |
| Found in RNA | Yes |
| Related nucleotides | 194 |
| Enzymes |
RlmC (Escherichia coli) RlmC (Pyrococcus abyssi) RlmCD (Bacillus subtilis) RlmD (Escherichia coli) RlmFO (Mycoplasma capricolum) TRMT2B (Mus musculus) Trm2 (Saccharomyces cerevisiae) TrmA (Escherichia coli) TrmFO (Bacillus subtilis) TrmFO (Thermus thermophilus) TrmU54 (Pyrococcus abyssi) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, tRNA |
Chemical information
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 6.154962408 |
| Energy [Eh]: | -950.028823543861 |
| HOMO [eV]: | -9.1953 |
| LUMO [eV]: | 0.8319 |
| Gap [eV]: | 10.0272 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cc1c(=O)[nH]c(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #0
Cc1c(O)nc(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #1 Cc1c(=O)nc(O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #2 |
| Tautomer image | Show Image |
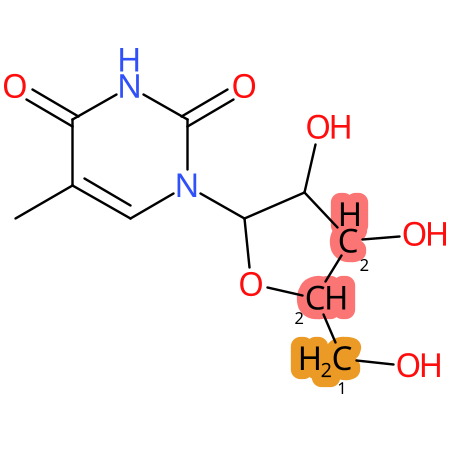
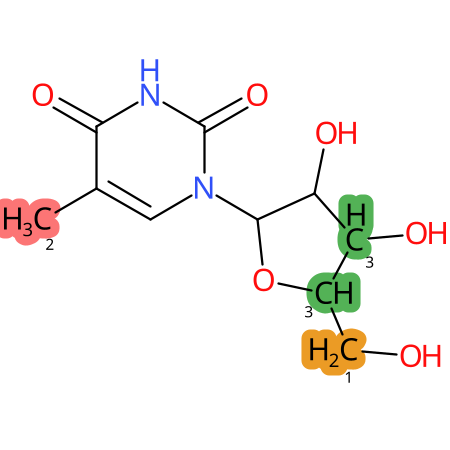
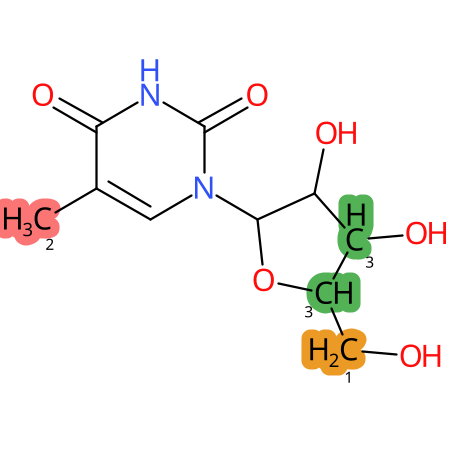
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 258.0852 |
| Average mass | 258.228 |
| [M+H]+ | 259.093 |
| Product ions | 127 |
| Normalized LC elution time * | 1 (Kellner 2014, Kellner 2014) |
| LC elution order/characteristics | co-eluting with G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Comments
Alternative name is ribosylthymidine.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic C |
Reactions producing 5-methyluridine
| Name |
|---|
| U:m5U |
Reactions starting from 5-methyluridine
| Name |
|---|
| m5U:m5Um |
| m5U:m5s2U |
Last modification of this entry: Sept. 15, 2025