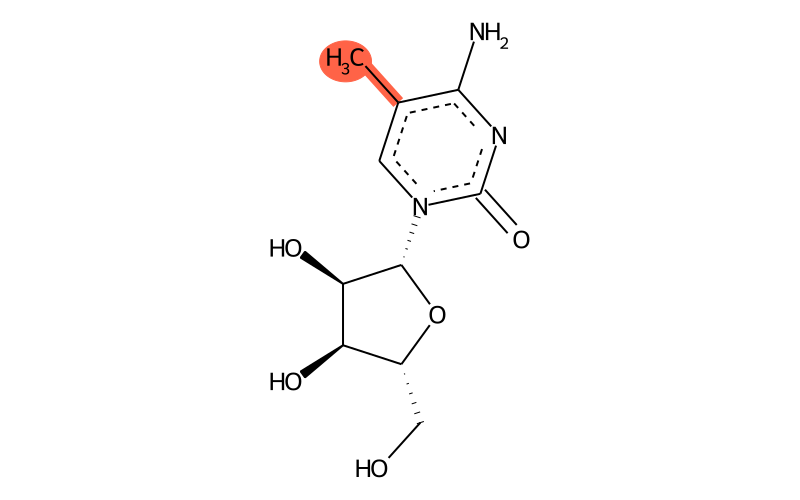
Summary
| Full name | 5-methylcytidine |
| IUPAC name | 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidin-2-one |
| Short name | m5C |
| MODOMICS code new | 2000000005C |
| MODOMICS code | 5C |
| Synonyms |
140M616
2140-61-6 21917-99-7 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2-dihydropyrimidin-2-one 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methylpyrimidin-2-one 4-amino-1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidin-2(1H)-one 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-5-methyl-pyrimidin-2-one 5-methyl-cytidine 5-Methylcytidine 5-Methylcytidine, 98% - 250MG 250mg 5-Methylcytidine, >=99% 5-Methyl-D-cytidine AC1L3P3S AC1Q6C23 AC-32357 Adenosine receptor agonist, 13 AJ-55618 AKOS015841917 AKOS015896918 AM83948 ANW-24421 AR-1G9002 BDBM200216 BRD-K65963527-001-01-8 C016568 CHEBI:20607 CHEMBL72086 CID92918 CS-0061684 CTK4E6675 Cytidine, 5-methyl- Cytidine,5-methyl- DS-8104 DTXSID401016977 EINECS 218-390-8 HY-113135 J-700100 M1931 m5C MFCD00006548 NS00049274 NSC 363933 Q238546 s6138 SB35398 SC-21988 SCHEMBL41572 ST24044891 TL9PB228DC TR-010033 UNII-TL9PB228DC ZAYHVCMSTBRABG-JXOAFFINSA-N ZINC6069529 |
| Nature of the modified residue | Natural |
| RNAMods code | ? |
| Residue unique ID | 18 |
| Found in RNA | Yes |
| Related nucleotides | 186 |
| Enzymes |
DnmA (Dictyostelium discoideum) Dnmt2 (Geobacter sulfurreducens) EfmM (Enterococcus faecium) NSUN1 (Homo sapiens) NSUN1 (Caenorhabditis elegans) NSUN4 (Mus musculus) Nop2 (Saccharomyces cerevisiae) Pmt1 (Schizosaccharomyces pombe) Rcm1 (Saccharomyces cerevisiae) RlmI (Escherichia coli) RlmO (Thermus thermophilus) RsmB (Escherichia coli) RsmB (Thermus thermophilus) RsmB (Haloferax volcanii) RsmF (Escherichia coli) RsmF (Thermus thermophilus) TRDMT1 (Homo sapiens) Trm4 (Saccharomyces cerevisiae) Trm4 (Homo sapiens) Trm4 (Pyrococcus abyssi) |
| Found in phylogeny | Eubacteria, Eukaryota |
| Found naturally in RNA types | mRNA, other, pre-tRNA, rRNA, tRNA |
Chemical information
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 9.288159408 |
| Energy [Eh]: | -930.1683481253 |
| HOMO [eV]: | -8.9222 |
| LUMO [eV]: | 0.9717 |
| Gap [eV]: | 9.8939 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cc1c(=N)[nH]c(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #0
Cc1c(N)nc(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #1 Cc1c(=N)nc(O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #2 |
| Tautomer image | Show Image |
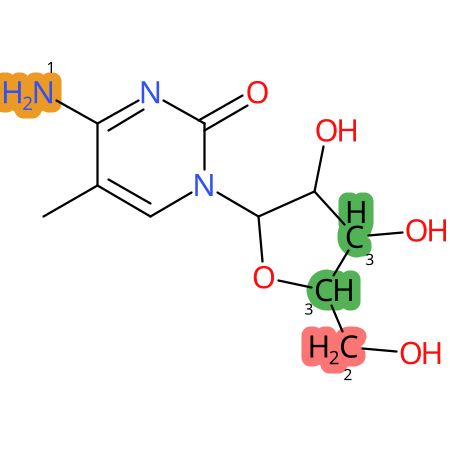
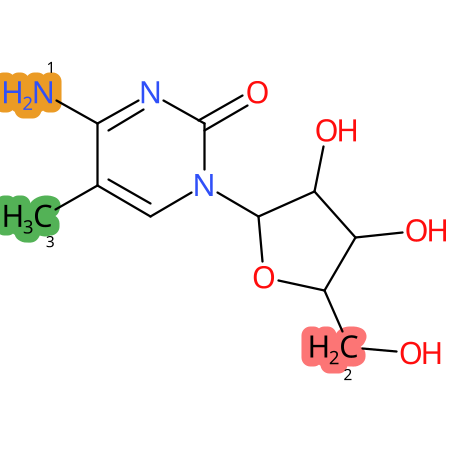
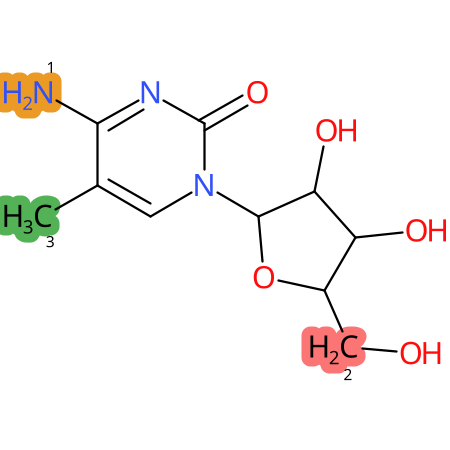
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 257.1012 |
| Average mass | 257.243 |
| [M+H]+ | 258.109 |
| Product ions | 126 |
| Normalized LC elution time * | 0,82 (Kellner 2014); 0,84 (Kellner 2014) |
| LC elution order/characteristics | between U and G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic C |
Reactions producing 5-methylcytidine
| Name |
|---|
| C:m5C |
Reactions starting from 5-methylcytidine
| Name |
|---|
| m5C:m5Cm |
| m5C:hm5C |
Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| A map of 5-methylcytosine residues in Trypanosoma brucei tRNA revealed by sodium bisulfite sequencing. | Militello KT, Chen LM, Ackerman SE, Mandarano AH, Valentine EL | Mol Biochem Parasitol | [details] | 24389163 | 10.1016 |
Last modification of this entry: Sept. 15, 2025