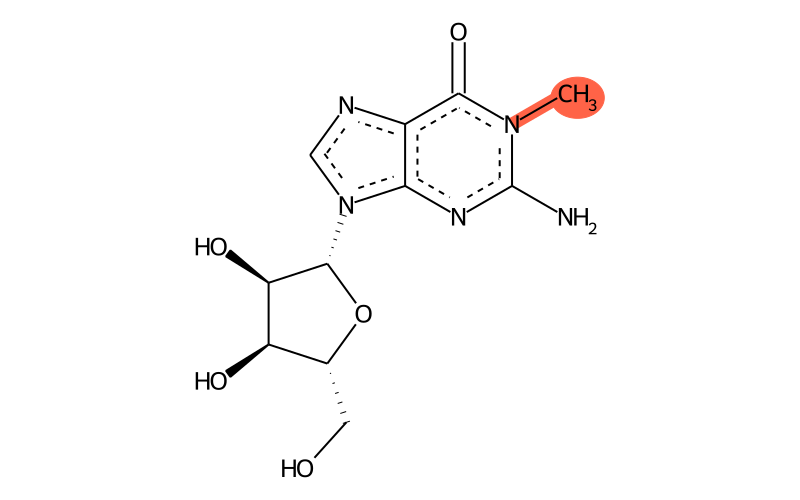
Summary
| Full name | 1-methylguanosine |
| IUPAC name | 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpurin-6-one |
| Short name | m1G |
| MODOMICS code new | 2000000001G |
| MODOMICS code | 1G |
| Synonyms |
1-methyl-guanosine
1-Methylguanosine 2140-65-0 21919-00-6 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methyl-6,9-dihydro-1H-purin-6-one 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpurin-6-one 2-amino-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-1-methyl-1,9-dihydro-6H-purin-6-one 2-Amino-9-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-1-methyl-1H-purin-6(9H)-one AC1L3WBB AC1Q3Y4I AC1Q6DXD AR-1J6689 CHEBI:19062 CHEMBL3246759 CID96373 CS-0061836 CTK1A4447 Guanosine, 1-methyl- Guanosine, 1-methyl- (8CI)(9CI) HY-113136 J-014033 m1g methylguanosine N1-Methylguanosine N/A N-methylguanosine NS00073958 NSC 70897 Q27109106 SCHEMBL62505 trmd protein ZINC5344125 |
| Nature of the modified residue | Natural |
| RNAMods code | K |
| Residue unique ID | 22 |
| Found in RNA | Yes |
| Related nucleotides | 214 |
| Enzymes |
AviRa (Streptomyces viridochromogenes) RlmA(I) (Escherichia coli) RlmA(II) (Streptomyces fradiae) TRM5 (Homo sapiens) TRMT10A (Homo sapiens) TRMT10B (Homo sapiens) TRMT10C (Homo sapiens) Taw22 (Pyrococcus abyssi) Trm10 (Saccharomyces cerevisiae) Trm10p (Thermococcus kodakaraensis) Trm5 (Saccharomyces cerevisiae) Trm5 (Haloferax volcanii) Trm5b (Methanocaldococcus jannaschii) Trm5b (Pyrococcus abyssi) TrmD (Escherichia coli) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, tRNA |
Chemical information
| Sum formula | C11H15N5O5 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 96373 |
| ChEBI ID | 19062 |
| CAS Registry Number | 2140-65-0 |
| Reaxys Registry Number | 570907 1226983 |
| SMILES | CN1C(=O)c2[n]c[n]([C@H]3[C@H](O)[C@H](O)[C@@H](CO)O3)c2N=C1N |
| logP | -2.0951 |
| TPSA | 148.65 |
| Number of atoms | 21 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 10 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C11H15N5O5/c1-15-9(20)5-8(14-11(15)12)16(3-13-5)10-7(19)6(18)4(2-17)21-10/h3-4,6-7,10,17-19H,2H2,1H3,(H2,12,14)/t4-,6-,7-,10-/m1/s1 |
| InChIKey | UTAIYTHAJQNQDW-KQYNXXCUSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
10228068
14849417 44426809 53801158 57336265 57394402 78991868 103830890 104418052 117476676 117590466 125317692 129416328 135053310 162499006 226443995 242627631 252400674 252410162 254770704 257685795 273528941 310278083 312226252 319441070 332868901 341161327 348275382 349717697 355073717 363671131 375987066 375987317 381990288 383850785 385663803 386500319 388623470 404622998 406854349 419580330 433776113 433776391 433776733 435910746 439442523 439576410 443531230 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 11.103810599 |
| Energy [Eh]: | -1077.72668863811 |
| HOMO [eV]: | -8.3688 |
| LUMO [eV]: | 1.519 |
| Gap [eV]: | 9.8878 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cn1c(=O)c2ncn(C3C(O)C(O)C(CO)O3)c2[nH]c1=N tautomer #0
Cn1c(=O)c2ncn(C3C(O)C(O)C(CO)O3)c2nc1N tautomer #1 Cn1c(O)c2ncn(C3C(O)C(O)C(CO)O3)c2nc1=N tautomer #2 CN1C(=O)C2N=CN(C3C(O)C(O)C(CO)O3)C2=NC1=N tautomer #3 |
| Tautomer image | Show Image |
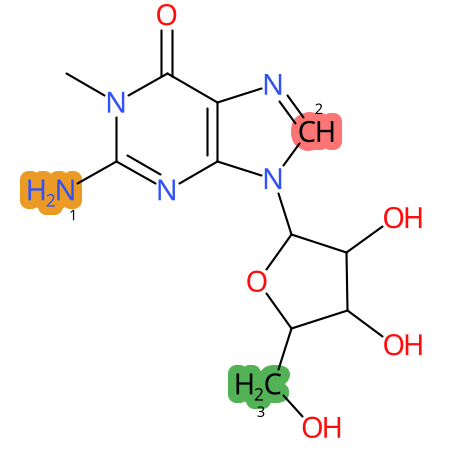
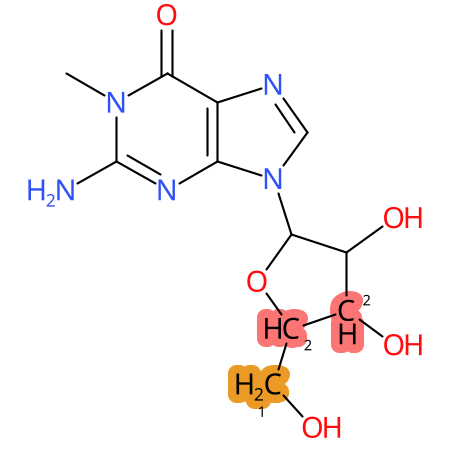
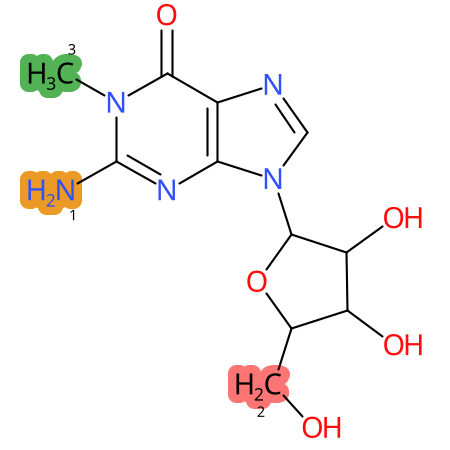
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 297.1073 |
| Average mass | 297.267 |
| [M+H]+ | 298.1151 |
| Product ions | 166 |
| Normalized LC elution time * | 1,23 (Kellner 2014); 1,28 (Kellner 2014) |
| LC elution order/characteristics | between G and A (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic N |
Reactions producing 1-methylguanosine
| Name |
|---|
| G:m1G |
Reactions starting from 1-methylguanosine
| Name |
|---|
| m1G:imG-14 |
| m1G:m1Gm |
Last modification of this entry: Sept. 15, 2025